
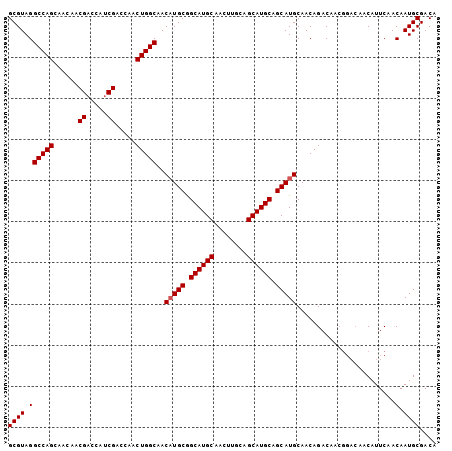
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,087,991 – 12,088,126 |
| Length | 135 |
| Max. P | 0.978686 |

| Location | 12,087,991 – 12,088,100 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12087991 109 + 23771897 UUUUCUGCCCAUCAAUUGC--UAGCAACAAAGCGCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGC--AUGCUGCAUGGAGCAUGCAGCAUGCAACAGAC ...((((............--...........(((((..(((((......((.....)).....)))))...)))))((--((((((((((...))))))))))))..)))). ( -39.00) >DroSec_CAF1 45322 111 + 1 UUUUCUGCCCAUCAAUUGCAAUAGCAACAAAGCGCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGC--AUGCAACUUGCAGCAUGCAGCAUGCAACAGAC ...((((........(((((...((.......(((((..(((((......((.....)).....)))))...)))))((--((((........)))))).)).))))))))). ( -32.00) >DroSim_CAF1 47579 111 + 1 UUUUCUGCCCAUCAAUUGCAAUAGCAACAAAGCGCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGC--AUGCAACUUGCAGCAUGCAGCAUGCAACAGAC ...((((........(((((...((.......(((((..(((((......((.....)).....)))))...)))))((--((((........)))))).)).))))))))). ( -32.00) >DroEre_CAF1 48028 106 + 1 UUUUCUGGCCAUCAAUUGC--CAGCAACAAAACGCGUAGGCCAGCAACAACGA---GCGACCAACUGGCAACUUGCAGC--AUGCAACUUGCAGCAUGCAGCAUGCAACAGAC ...((((........((((--((((........))((.((...((........---))..)).))))))))..(((.((--((((........)))))).))).....)))). ( -29.80) >DroYak_CAF1 46730 108 + 1 UUUGCUGGCCAUCAAUUGG--CAGCAACAAAACGCGUAGGCCAGCAACAACGA---GCGACCAACUGGCAACAUGCAGCACAUGCAACUUGCAGCAUGCAGCAUGCAACAGAC .(((((((((.((....))--..((........))...)))))))))......---........(((....(((((.(((..(((.....)))...))).)))))...))).. ( -33.70) >consensus UUUUCUGCCCAUCAAUUGC__UAGCAACAAAGCGCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGC__AUGCAACUUGCAGCAUGCAGCAUGCAACAGAC ...((((........((((....))))......((((..(((((......((.....)).....))))).....((.((..((((........)))))).))))))..)))). (-22.70 = -23.10 + 0.40)


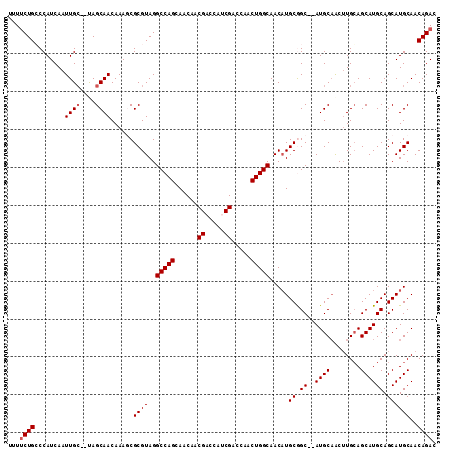
| Location | 12,087,991 – 12,088,100 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -29.20 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12087991 109 - 23771897 GUCUGUUGCAUGCUGCAUGCUCCAUGCAGCAU--GCCGCAUGUUGCCAGUUGGUCGAUGGUCGUUGUUGCUGGCCUACGCGCUUUGUUGCUA--GCAAUUGAUGGGCAGAAAA ...(((.((((((((((((...))))))))))--)).)))..(((((.((..((((((....)))).(((((((..(((.....))).))))--)))))..)).))))).... ( -43.90) >DroSec_CAF1 45322 111 - 1 GUCUGUUGCAUGCUGCAUGCUGCAAGUUGCAU--GCCGCAUGUUGCCAGUUGGUCGAUGGUCGUUGUUGCUGGCCUACGCGCUUUGUUGCUAUUGCAAUUGAUGGGCAGAAAA .((((((((((((.((((((........))))--)).)))))).((((((....((((....))))..)))))).....((.((.(((((....))))).))))))))))... ( -40.70) >DroSim_CAF1 47579 111 - 1 GUCUGUUGCAUGCUGCAUGCUGCAAGUUGCAU--GCCGCAUGUUGCCAGUUGGUCGAUGGUCGUUGUUGCUGGCCUACGCGCUUUGUUGCUAUUGCAAUUGAUGGGCAGAAAA .((((((((((((.((((((........))))--)).)))))).((((((....((((....))))..)))))).....((.((.(((((....))))).))))))))))... ( -40.70) >DroEre_CAF1 48028 106 - 1 GUCUGUUGCAUGCUGCAUGCUGCAAGUUGCAU--GCUGCAAGUUGCCAGUUGGUCGC---UCGUUGUUGCUGGCCUACGCGUUUUGUUGCUG--GCAAUUGAUGGCCAGAAAA .((((..((.(((.((((((........))))--)).)))((((((((((.((((((---........)).)))).))(((......)))))--))))))....))))))... ( -39.80) >DroYak_CAF1 46730 108 - 1 GUCUGUUGCAUGCUGCAUGCUGCAAGUUGCAUGUGCUGCAUGUUGCCAGUUGGUCGC---UCGUUGUUGCUGGCCUACGCGUUUUGUUGCUG--CCAAUUGAUGGCCAGCAAA ....((.((((((.(((((.(((.....)))))))).)))))).))..((.((((((---........)).)))).))........((((((--(((.....))).)))))). ( -37.80) >consensus GUCUGUUGCAUGCUGCAUGCUGCAAGUUGCAU__GCCGCAUGUUGCCAGUUGGUCGAUGGUCGUUGUUGCUGGCCUACGCGCUUUGUUGCUA__GCAAUUGAUGGGCAGAAAA (((((((((((((.((((((........))))..)).))))((.((((((....((.....)).....))))))..)))).....(((((....))))).)))))))...... (-29.20 = -30.00 + 0.80)


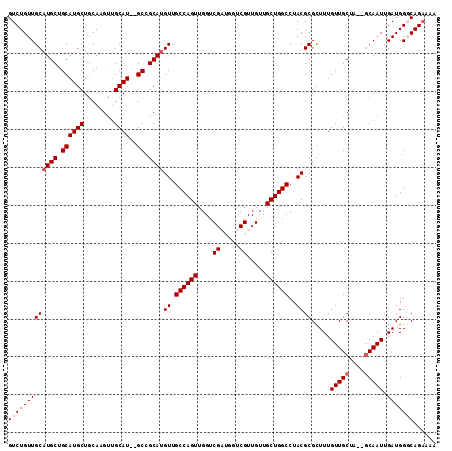
| Location | 12,088,022 – 12,088,126 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.46 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -27.77 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
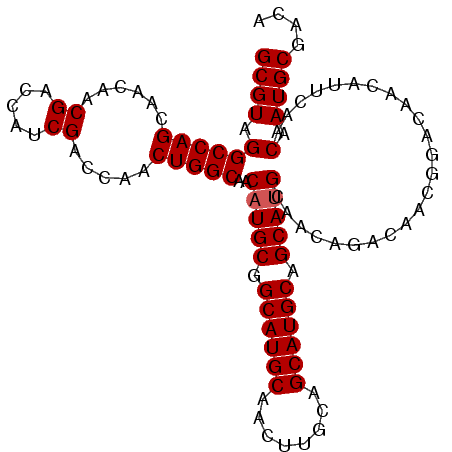
>3L_DroMel_CAF1 12088022 104 + 23771897 GCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGCAUGCUGCAUGGAGCAUGCAGCAUGCAACAGACAACGGACAACAUUCAACAAUGCGACA ((((.((((((......((.....)).....)))))....((..((((((((((((...))))))))))))..))...................).)))).... ( -35.10) >DroSec_CAF1 45355 104 + 1 GCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGCAUGCAACUUGCAGCAUGCAGCAUGCAACAGACAACGGACAACAUUCGACAAUGCGACA ((((.((((((......((.....)).....)))))..(((((.((((((........)))))).)))))..........((((.....)))).).)))).... ( -30.50) >DroEre_CAF1 48059 94 + 1 GCGUAGGCCAGCAACAACGA---GCGACCAACUGGCAACUUGCAGCAUGCAACUUGCAGCAUGCAGCAUGCAACAGAC-------AACAUUCAACAAUGCGACA ((((.((...((........---))..))..(((((((.((((.....)))).)))).(((((...)))))..)))..-------...........)))).... ( -24.00) >consensus GCGUAGGCCAGCAACAACGACCAUCGACCAACUGGCAACAUGCGGCAUGCAACUUGCAGCAUGCAGCAUGCAACAGACAACGGACAACAUUCAACAAUGCGACA ((((.((((((......((.....)).....)))))..(((((.((((((........)))))).)))))........................).)))).... (-27.77 = -28.10 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:14 2006