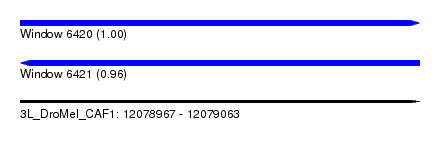
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,078,967 – 12,079,063 |
| Length | 96 |
| Max. P | 0.998393 |

| Location | 12,078,967 – 12,079,063 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -15.51 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12078967 96 + 23771897 GCGGCCCACA--CACAAUCUUUCAUUAUAACUAAUGAACGUUUCGCGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA (((((.....--..(((((.(((((((....)))))))((.....))..).))))..((((((.......)))))).............))))).... ( -16.80) >DroSec_CAF1 36528 96 + 1 UAUGCCCACA--CACAAUCUUUCAUUAUAACUAAUGAACGUUUCGCGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA ..........--........(((((((....)))))))......((((((..(((.(((((((.......)))))).....).)))..)))))).... ( -15.00) >DroSim_CAF1 38298 96 + 1 UAUGCCCACA--CACAAUCUUUCAUUAUAACUAAUGAACGUUUCGCGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA ..........--........(((((((....)))))))......((((((..(((.(((((((.......)))))).....).)))..)))))).... ( -15.00) >DroEre_CAF1 38981 96 + 1 GUGGCCCACA--CACAAUCUUUCAUUAUAACUAAUGAACGUUUCGUGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA (((((((((.--......(.(((((((....))))))).)....)))).........((((((.......)))))).............))))).... ( -16.50) >DroYak_CAF1 37292 98 + 1 CUGGCCCACACACACAAUCUUUCAUUAUAACUAAUGAACGUUUCGUGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA ....................(((((((....)))))))......((((((..(((.(((((((.......)))))).....).)))..)))))).... ( -13.10) >DroAna_CAF1 35694 96 + 1 GAGGCCCACA--CACAAUAUUUCAUUAUAACUAAUGAACGUUUCGCGGCACAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA (((((.....--........(((((((....))))))).)))))((((((..(((.(((((((.......)))))).....).)))..)))))).... ( -16.64) >consensus GAGGCCCACA__CACAAUCUUUCAUUAUAACUAAUGAACGUUUCGCGGCGCAUUGACAAUUAAUCAAAAUUUAAUUAAAUAGACAAAAUGCCGCAUUA ....................(((((((....)))))))......((((((..(((.(((((((.......)))))).....).)))..)))))).... (-14.84 = -14.48 + -0.36)

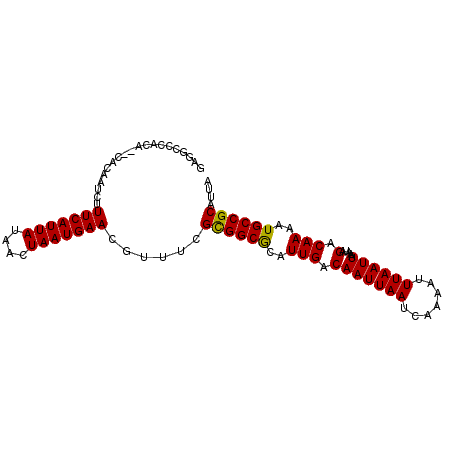
| Location | 12,078,967 – 12,079,063 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12078967 96 - 23771897 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCGCGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUG--UGUGGGCCGC ....(((((...(((.(...(((((((.....)))))))..).)))..((((((((....(((((((....)))))))...)))))--.))).))))) ( -25.90) >DroSec_CAF1 36528 96 - 1 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCGCGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUG--UGUGGGCAUA ...((((((...(((.(...(((((((.....)))))))..).)))...))))))....((((((....((((((.....))))))--.))))))... ( -23.00) >DroSim_CAF1 38298 96 - 1 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCGCGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUG--UGUGGGCAUA ...((((((...(((.(...(((((((.....)))))))..).)))...))))))....((((((....((((((.....))))))--.))))))... ( -23.00) >DroEre_CAF1 38981 96 - 1 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCACGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUG--UGUGGGCCAC ......(((...(((.(...(((((((.....)))))))..).)))..((((((((....(((((((....)))))))...)))))--.))).))).. ( -22.50) >DroYak_CAF1 37292 98 - 1 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCACGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUGUGUGUGGGCCAG ......(((...(((.(...(((((((.....)))))))..).))).(((((((((....(((((((....)))))))...))))).))))..))).. ( -23.90) >DroAna_CAF1 35694 96 - 1 UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGUGCCGCGAAACGUUCAUUAGUUAUAAUGAAAUAUUGUG--UGUGGGCCUC ...((((((((.(((.(...(((((((.....)))))))..).))).))))))))....((((((....(((((((...)))))))--.))))))... ( -25.60) >consensus UAAUGCGGCAUUUUGUCUAUUUAAUUAAAUUUUGAUUAAUUGUCAAUGCGCCGCGAAACGUUCAUUAGUUAUAAUGAAAGAUUGUG__UGUGGGCCUC ...((((((...(((.(...(((((((.....)))))))..).)))...))))))....((((((....((((((.....))))))...))))))... (-20.65 = -20.98 + 0.33)

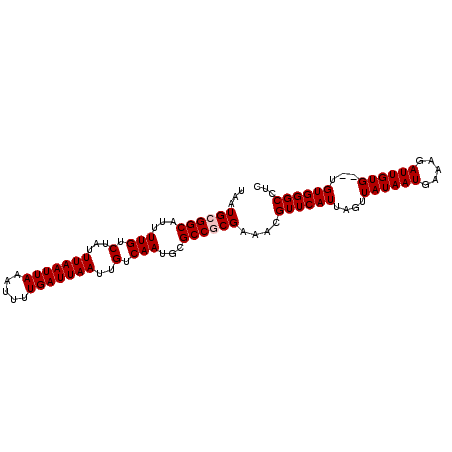
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:10 2006