| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,036,037 – 12,036,152 |
| Length | 115 |
| Max. P | 0.589752 |

| Location | 12,036,037 – 12,036,152 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -32.55 |
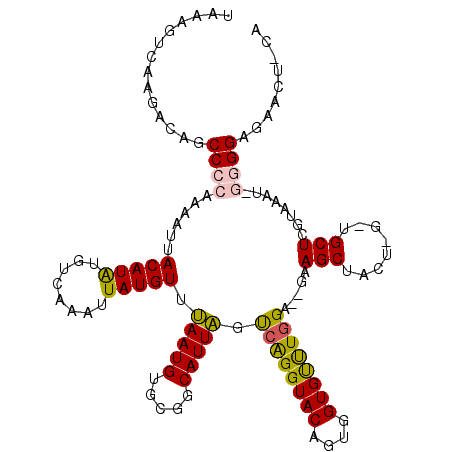
| Consensus MFE | -17.57 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
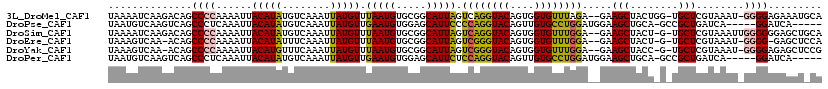
>3L_DroMel_CAF1 12036037 115 - 23771897 UAAAAUCAAGACAGCCCCAAAAUUACAUAUGUCAAAUUAUGUUUAAUGUGCGGCAUUAGUCAGGUACAGUGGUGUUUAGA--GAAGCUACUGG-UGCUCGUAAAU-GGGGAGAAAUGCA ..............(((((...((((.............((.((((((.....)))))).))(((((((((((.((....--)).)))))).)-)))).)))).)-))))......... ( -25.70) >DroPse_CAF1 51493 108 - 1 UAAUGUCAAGUCAGCCCUCAAAUUACAUAUGUCAAAUUAUGUUGAAUGUGGAGCAUUCCCCAGGUACAGUUGUGCCUGGAUGGAAGCUGCA-GCCGCUGAUCA-----GGAUCA----- ....(((..((((((((.((....(((((........)))))....)).))(((.((((((((((((....))))))))..)))))))...-...))))))..-----.)))..----- ( -36.80) >DroSim_CAF1 53130 115 - 1 UAAAAUCAAGACAGCCCCAAAAUUACAUAUGUCAAAUUAUGUUUAAUGUGCGGCAUUAGUCAGGUACAGUGGUGUUUGGA--GAAGCUACU-G-UGCUCGUAAAUUGGGGGGAGCUGCA .........(.(((((((......(((((........)))))(((((.((((((....))).(((((((((((.((....--)).))))))-)-)))).))).))))).))).))))). ( -33.70) >DroEre_CAF1 55885 112 - 1 UAAAGUCAA-ACAGCCCCAAAAUUACAUAUUUCAAAUUAUGUUUAAUGUGCGGCAUUAGUCGGGUACAGUGGUGUUUGGA--GAAGCUACU-G-UGCUCGUAAAU-GGGG-GAGCUCCA .........-....(((((.....(((((........)))))((((((.....)))))).(((((((((((((.((....--)).))))))-)-))))))....)-))))-........ ( -33.00) >DroYak_CAF1 54756 113 - 1 UAAAGUCAA-ACAGCCCCAAAAUUACAUGUUUCAAAUUAUGUUUAAUGUGCGGCAUUAGUCGGGUACAGUGGUGUUUGGA--GAAGCUACC-G-UGCUCGUAAAU-GGGGAGAGCUCCG .........-....(((((...((((((((..((.((((....)))).))..)))).....((((((.(((((.((....--)).))))).-)-))))))))).)-))))......... ( -29.00) >DroPer_CAF1 48396 108 - 1 UAAUGUCAAGUCAGCCCUCAAAUUACAUAUGUCAAAUUAUGUUGAAUGUGGAGCAUUCUCCAGGUACAGUUGUGCCUGGAUGGAAGCUGCA-GCCGCUGAUCA-----GGAUCA----- ....(((..((((((((.((....(((((........)))))....)).))(((.((((((((((((....))))))))).))).)))...-...))))))..-----.)))..----- ( -37.10) >consensus UAAAGUCAAGACAGCCCCAAAAUUACAUAUGUCAAAUUAUGUUUAAUGUGCGGCAUUAGUCAGGUACAGUGGUGUUUGGA__GAAGCUACU_G_UGCUCGUAAAU_GGGGAGAACU_CA ..............((((......(((((........))))).(((((.....))))).((((((((....)))))))).....(((........)))........))))......... (-17.57 = -16.93 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:01 2006