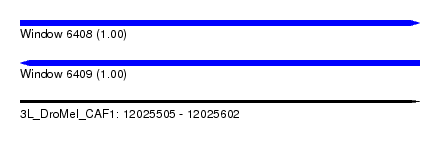
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,025,505 – 12,025,602 |
| Length | 97 |
| Max. P | 0.999985 |

| Location | 12,025,505 – 12,025,602 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.66 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 5.37 |
| SVM RNA-class probability | 0.999985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
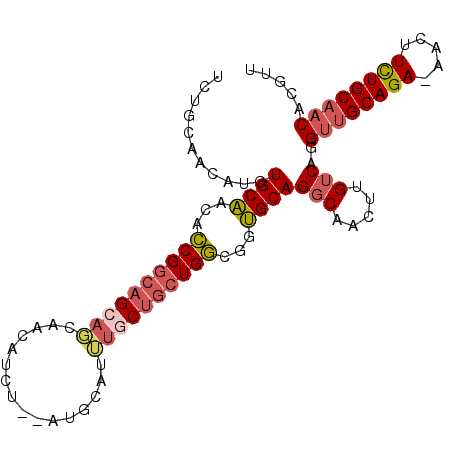
>3L_DroMel_CAF1 12025505 97 + 23771897 UCUGCAACAUGUGCAACACCGGCAGCAGCAACAUCC--AUGCAUUUACUGCUGGCGGUGCAGGCAACUUGUCAGGUUGCAGA-AACUUCUGCAACACGUU .((((((....(((..(((((.((((((........--.........)))))).)))))...)))..))).)))((((((((-....))))))))..... ( -38.73) >DroSim_CAF1 42526 89 + 1 UCUGCAACAUGUGCAACACCGGCAGCAGCAACAUUU----------GCUGCUGGCGGUGCAGGCAACUUGUCAGGUUGCAGA-AACUUUUGCAACACGUU .((((((....(((..(((((.((((((((.....)----------))))))).)))))...)))..))).)))((((((((-....))))))))..... ( -42.30) >DroEre_CAF1 45587 95 + 1 UCUGCAACAACUGCUUCACCGGCAGCAGCAACAUC---AUGCAUUUGCUGCUGGUG-AGCAGGCGACUUGUCAGGUUGCAGA-AACUUCUGCAACACGUU .((((((...(((((.((((((((((((((.....---.)))...)))))))))))-))))).....))).)))((((((((-....))))))))..... ( -46.10) >DroWil_CAF1 49146 98 + 1 AAGGAAAGUUGUGCAA--GCGACAGCAUUUAAGUGUGUGUGUGUGUCCCUCUGUGUGUGCAGUCAAGUUGUCAGGCGGCAGAAAACUUCUGCAACAUGUU ..(((..(((((....--))))).((((......)))).......)))....((((.(((((.....(((((....))))).......)))))))))... ( -21.90) >consensus UCUGCAACAUGUGCAACACCGGCAGCAGCAACAUCU__AUGCAUUUGCUGCUGGCGGUGCAGGCAACUUGUCAGGUUGCAGA_AACUUCUGCAACACGUU ...........((((...((((((((((................))))))))))...))))(((.....)))..((((((((.....))))))))..... (-23.35 = -24.66 + 1.31)

| Location | 12,025,505 – 12,025,602 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
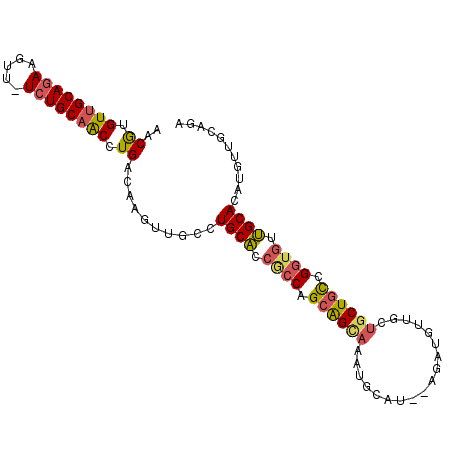
>3L_DroMel_CAF1 12025505 97 - 23771897 AACGUGUUGCAGAAGUU-UCUGCAACCUGACAAGUUGCCUGCACCGCCAGCAGUAAAUGCAU--GGAUGUUGCUGCUGCCGGUGUUGCACAUGUUGCAGA .....((((((((....-))))))))((((((.((.((..((((((.((((((((...(((.--...))))))))))).)))))).)))).)))..))). ( -40.50) >DroSim_CAF1 42526 89 - 1 AACGUGUUGCAAAAGUU-UCUGCAACCUGACAAGUUGCCUGCACCGCCAGCAGC----------AAAUGUUGCUGCUGCCGGUGUUGCACAUGUUGCAGA (((((((.(((......-...(((((.......)))))..((((((.(((((((----------(.....)))))))).))))))))))))))))..... ( -38.50) >DroEre_CAF1 45587 95 - 1 AACGUGUUGCAGAAGUU-UCUGCAACCUGACAAGUCGCCUGCU-CACCAGCAGCAAAUGCAU---GAUGUUGCUGCUGCCGGUGAAGCAGUUGUUGCAGA .....((((((((....-))))))))(((.(((..((.(((((-((((.((((((...(((.---.....))))))))).)))).))))).)))))))). ( -42.00) >DroWil_CAF1 49146 98 - 1 AACAUGUUGCAGAAGUUUUCUGCCGCCUGACAACUUGACUGCACACACAGAGGGACACACACACACACUUAAAUGCUGUCGC--UUGCACAACUUUCCUU ....(((((((((((((.((........)).)))))..))))).)))..(((.((((((..............)).)))).)--)).............. ( -17.34) >consensus AACGUGUUGCAGAAGUU_UCUGCAACCUGACAAGUUGCCUGCACCGCCAGCAGCAAAUGCAU__AGAUGUUGCUGCUGCCGGUGUUGCACAUGUUGCAGA ..((.((((((((.....)))))))).))..........((((.((((.((((((..................)))))).)))).))))........... (-23.25 = -23.87 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:58 2006