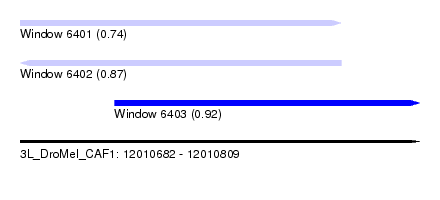
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,010,682 – 12,010,809 |
| Length | 127 |
| Max. P | 0.918979 |

| Location | 12,010,682 – 12,010,784 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12010682 102 + 23771897 CUUGGAGCAUUAGUCUUAUGCAUAAUCGGUGGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGACUAACUA .........((((((((((((((....((.....))...))))))))....((((((((....))))))))......................))))))... ( -23.00) >DroSec_CAF1 28768 102 + 1 CUUGGAGCAUUAGUCUUAUGCAUAAUCGGUGGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGACUAACUA .........((((((((((((((....((.....))...))))))))....((((((((....))))))))......................))))))... ( -23.00) >DroSim_CAF1 28005 102 + 1 CUUGGAGCAUUAGUCUUAUGCAUAAUCGGUGGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCGUAUAUCUGCCUAACUA .((((.(((((((.(((((.........))))).)))..))))........((((((((....))))))))))))........................... ( -22.30) >DroEre_CAF1 29453 102 + 1 CUUGGAGCAUUAGUCUUAUACAUAAUCGGUGGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGCCUAACUA .((((.(((((((.(((((.........))))).)))..))))........((((((((....))))))))))))........................... ( -22.60) >DroYak_CAF1 28703 102 + 1 CUUGGAGCAUUAGUCUUAUGCAUAAUCGGUGGGCCUCAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUUUAUCUGCCUAACUA .((((.(((......((((((((....((....))....))))))))....((((((((....)))))))).....................)))))))... ( -24.90) >DroAna_CAF1 27020 81 + 1 CUUGGAUCAUUAGUUUUAAACUUAAUCAGUUG-ACUAAAAUGCAUAAUUAUGCAUUUGAAUUCCUAUAUGCCUUUAAGCUUU-------------------- ...((((((((((((...((((.....)))))-)))))(((((((....))))))))))..))).....((......))...-------------------- ( -12.90) >consensus CUUGGAGCAUUAGUCUUAUGCAUAAUCGGUGGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGCCUAACUA .((((.(((((((.(((((.........))))).)))..))))........((((((((....))))))))))))........................... (-16.62 = -17.57 + 0.95)


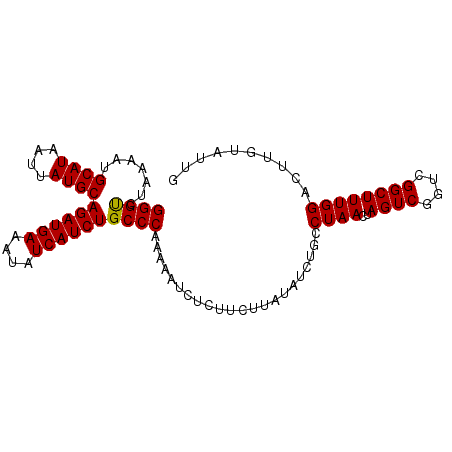
| Location | 12,010,682 – 12,010,784 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12010682 102 - 23771897 UAGUUAGUCAGAUAUAAGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUUAGACCCACCGAUUAUGCAUAAGACUAAUGCUCCAAG ..(((((((.....((((((....)))))).((((((((....))))))))....((((((((....(......)....)))))))))))))))........ ( -28.80) >DroSec_CAF1 28768 102 - 1 UAGUUAGUCAGAUAUAAGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUUAGACCCACCGAUUAUGCAUAAGACUAAUGCUCCAAG ..(((((((.....((((((....)))))).((((((((....))))))))....((((((((....(......)....)))))))))))))))........ ( -28.80) >DroSim_CAF1 28005 102 - 1 UAGUUAGGCAGAUAUACGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUUAGACCCACCGAUUAUGCAUAAGACUAAUGCUCCAAG ....................(((....((((((((((((....))))))))....((((((((....(......)....))))))))..))))..))).... ( -25.10) >DroEre_CAF1 29453 102 - 1 UAGUUAGGCAGAUAUAAGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUUAGACCCACCGAUUAUGUAUAAGACUAAUGCUCCAAG ....................(((....((((((((((((....))))))))....((((((((....(......)....))))))))..))))..))).... ( -23.10) >DroYak_CAF1 28703 102 - 1 UAGUUAGGCAGAUAAAAGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUGAGGCCCACCGAUUAUGCAUAAGACUAAUGCUCCAAG ....................(((....((((((((((((....))))))))....((((((((....((....))....))))))))..))))..))).... ( -27.80) >DroAna_CAF1 27020 81 - 1 --------------------AAAGCUUAAAGGCAUAUAGGAAUUCAAAUGCAUAAUUAUGCAUUUUAGU-CAACUGAUUAAGUUUAAAACUAAUGAUCCAAG --------------------...(((....))).....(((..((((((((((....)))))))(((((-(....))))))............))))))... ( -15.50) >consensus UAGUUAGGCAGAUAUAAGAAGAGAUUUUUGGGCAGAUGAUAUUUCAUCUGCAUAAUUAUGCAUUUUAGACCCACCGAUUAUGCAUAAGACUAAUGCUCCAAG ....................(((....((((((((((((....))))))))....((((((((....(......)....))))))))..))))..))).... (-19.68 = -20.10 + 0.42)


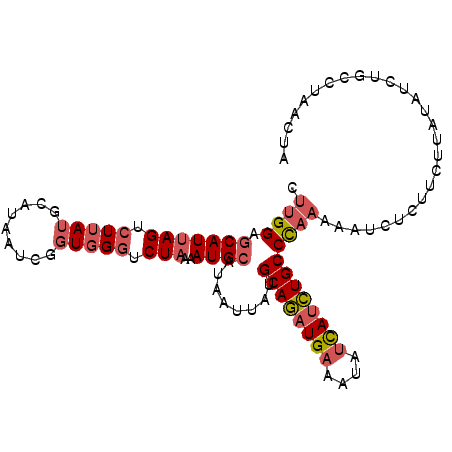
| Location | 12,010,712 – 12,010,809 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -19.04 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12010712 97 + 23771897 GGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGACUAACUAGUCGCUCGGCUUUGGAUUUGUAUUG ........((((((.......((((((((....))))))))(((((.................((((....))))((...)))))))...)))))). ( -21.80) >DroSec_CAF1 28798 97 + 1 GGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGACUAACUAGUCGGUCGGCUUUGGACUUGUAUUG (((((((((..((........((((((((....))))))))....................((((((....))))))...)))))))))))...... ( -26.40) >DroSim_CAF1 28035 97 + 1 GGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCGUAUAUCUGCCUAACUAGUCGGUCGGCUUUGGACUUGUAUUG (((((((((............((((((((....))))))))......................(((..(((....))).))))))))))))...... ( -22.80) >DroEre_CAF1 29483 96 + 1 GGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGCCUAACUAGUCGGUCGGCUUUGGACUUGU-AUU (((((((((............((((((((....))))))))......................(((..(((....))).))))))))))))..-... ( -22.80) >DroYak_CAF1 28733 97 + 1 GGGCCUCAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUUUAUCUGCCUAACUAGUCGGUUGGCUUUGGAGUUGUUAUC ((((.......((((....))))((((((....))))))))))...(((..((((........(((.((((....)))))))...))))..)))... ( -24.50) >consensus GGGUCUAAAAUGCAUAAUUAUGCAGAUGAAAUAUCAUCUGCCCAAAAAUCUCUUCUUAUAUCUGCCUAACUAGUCGGUCGGCUUUGGACUUGUAUUG ((((.......((((....))))((((((....))))))))))......................((((..((((....)))))))).......... (-19.04 = -18.88 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:52 2006