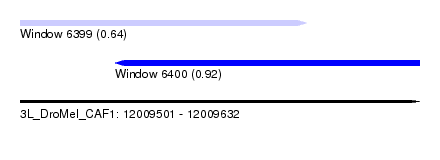
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,009,501 – 12,009,632 |
| Length | 131 |
| Max. P | 0.923424 |

| Location | 12,009,501 – 12,009,595 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
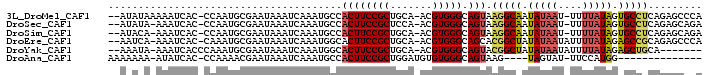
>3L_DroMel_CAF1 12009501 94 + 23771897 --AUAUAAAAAUCAC-CCAAUGCGAAUAAAUCAAAUGCCACUUCCGCUGCA-ACGUGGGCAGUAAGGCAAUAUAAU-UUUUAUAGUGCCUCAGAGCCCA --.............-....(((((.....))....((.......)).)))-...(((((..(.(((((.((((..-...)))).))))).)..))))) ( -18.30) >DroSec_CAF1 27638 93 + 1 --AUAUA-AAAUCAC-CCAAUGCGAAUAAAUCAAAUGCCACUUCCGCUCCA-ACGUGGGCAGUAAGGCAAUAUAAU-UUUUAUAGUGCCUCAGAGCAGA --.....-.......-....(((................((((((((....-..))))).))).(((((.((((..-...)))).)))))....))).. ( -17.90) >DroSim_CAF1 26852 93 + 1 --AUACA-AAAUCAC-CCAAUGCGAAUAAAUCAAAUGCCACUUCCGCUGCA-ACGUGGGCAGUAAGGCAAUAUAAU-UUUUAUAGUGCCUCAGAGCAGA --.....-.......-....(((................((((((((....-..))))).))).(((((.((((..-...)))).)))))....))).. ( -17.90) >DroEre_CAF1 28237 94 + 1 --AAUCA-AAAUCAC-CAAAUGCGAAUAAAUCAAAUGGCACUUCCGCUGCA-ACGUGGGCAGCACGGCUAUAUAAUAUUUUAUAGAGCCGCAGAGCCCA --.....-.......-....................(((......(((((.-......))))).(((((.(((((....))))).)))))....))).. ( -23.80) >DroYak_CAF1 27525 88 + 1 --AAAUA-AAAUCACCCAAAUGCGAAUAAAUCAAAUGGCACUUCCGCUGCA-ACGUGGGCAGUACGGCUAUAUAAUAUUUUAUAGAGCUGCA------- --.....-......((((..(((((.....))....(((......))))))-...)))).....(((((.(((((....))))).)))))..------- ( -19.00) >DroAna_CAF1 26202 78 + 1 AAAAAAA-AUAUCAC-CCAAAACGAAUAAAUCAAAUGCCACUUCCGCUGGAUGUGUGGGCAGUAAG----UAGUAU-UUCCAUGG-------------- .......-.......-(((....(((((.......((((((.(((...)))...)).)))).....----...)))-))...)))-------------- ( -10.86) >consensus __AAAUA_AAAUCAC_CCAAUGCGAAUAAAUCAAAUGCCACUUCCGCUGCA_ACGUGGGCAGUAAGGCAAUAUAAU_UUUUAUAGUGCCUCAGAGC__A .......................................((((((((.......))))).))).(((((.(((((....))))).)))))......... (-12.46 = -12.67 + 0.21)


| Location | 12,009,532 – 12,009,632 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -17.36 |
| Energy contribution | -18.57 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
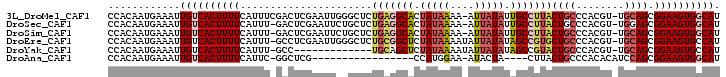
>3L_DroMel_CAF1 12009532 100 - 23771897 CCACAAUGAAAUUGUCACUUUUCAUUUCGACUCGAAUUGGGCUCUGAGGCACUAUAAAA-AUUAUAUUGCCUUACUGCCCACGU-UGCAGCGGAAGUGGCAU ((((..(((((........)))))(((((.(((((..(((((..(((((((.((((...-..)))).)))))))..)))))..)-)).)))))))))))... ( -36.10) >DroSec_CAF1 27668 99 - 1 CCACAAUGAAAUUGUCACUUUUCAUUU-GACUCGAAUUCUGCUCUGAGGCACUAUAAAA-AUUAUAUUGCCUUACUGCCCACGU-UGGAGCGGAAGUGGCAU (((((((((((........))))))).-........(((((((((((((((.((((...-..)))).)))))))(.((....))-.)))))))))))))... ( -29.40) >DroSim_CAF1 26882 99 - 1 CCACAAUGAAAUUGUCACUUUUCAUUU-GACUCGAAUUCUGCUCUGAGGCACUAUAAAA-AUUAUAUUGCCUUACUGCCCACGU-UGCAGCGGAAGUGGCAU (((((((((((........))))))).-........((((((..(((((((.((((...-..)))).))))))).(((......-.)))))))))))))... ( -27.40) >DroEre_CAF1 28267 100 - 1 CCACAAUGAAAUUGUCACUUUUCAUUU-GCCUCGAAUUGGGCUCUGCGGCUCUAUAAAAUAUUAUAUAGCCGUGCUGCCCACGU-UGCAGCGGAAGUGCCAU ............((.(((((((...((-((..((...(((((..(((((((.(((((....))))).)))))))..))))))).-.)))).))))))).)). ( -33.80) >DroYak_CAF1 27556 87 - 1 CCACAAUGAAAUUGUCACUUUUCAUUU-GCC-------------UGCAGCUCUAUAAAAUAUUAUAUAGCCGUACUGCCCACGU-UGCAGCGGAAGUGCCAU .(((..(((((........)))))(((-((.-------------(((((((.(((((....))))).)))(((.......))).-))))))))).))).... ( -17.50) >DroAna_CAF1 26234 79 - 1 CCACAAUGAAAUUGUCACUUUUCAUUC-GGCUCG-----------------CCAUGGAA-AUACUA----CUUACUGCCCACACAUCCAGCGGAAGUGGCAU (((.(((((((........))))))).-((....-----------------)).)))..-...(((----(((.((((...........))))))))))... ( -17.10) >consensus CCACAAUGAAAUUGUCACUUUUCAUUU_GACUCGAAUU__GCUCUGAGGCACUAUAAAA_AUUAUAUUGCCUUACUGCCCACGU_UGCAGCGGAAGUGGCAU ............((((((((((......................(((((((.(((((....))))).)))))))((((........)))).)))))))))). (-17.36 = -18.57 + 1.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:49 2006