| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,002,858 – 12,002,964 |
| Length | 106 |
| Max. P | 0.618015 |

| Location | 12,002,858 – 12,002,964 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -10.74 |
| Energy contribution | -12.97 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12002858 106 + 23771897 -UAAAGUUGGCAUCCAUAAAA----AAGCGAAACAG--UUUAAAGCGUUUCGAAAUGGUAAUGAGCAAAUUACGAGAAGCCCUUCAAAGGGCUU-CACAACCAGGUGUUUAUUU-- -..((((..(((((.......----...((((((.(--(.....))))))))...(((((((......)))))..((((((((....)))))))-).))....)))))..))))-- ( -29.10) >DroPse_CAF1 21937 102 + 1 -UAUCGAGUGUAAGUGUAAAUGUCUUCGAGAAACAGGGCUAAAAGCUG------GCGUUAAUGGGGAAAUUA-----AGUACUA-GAAGGGUU-CCACAGGCAGGUGAUAUUUCAA -(((((..(((..(((.((...(((((.((......(((.....))).------((.(((((......))))-----))).)).-))))).))-.)))..)))..)))))...... ( -18.90) >DroSec_CAF1 21129 107 + 1 -UAAAGUGAGCAACCAUAAAA----ACGCGAAACAG--UUUAAAGCGUUUCGAAAUGGUAAUGAGCAAAUUACGAGAAGCCCUUUAAAGGGUUUCCACAAGCAGGUGUUUAUUU-- -..(((((((((.((......----...((((((.(--(.....))))))))...(((((((......)))))(.((((((((....))))))))).))....)))))))))))-- ( -29.80) >DroSim_CAF1 20351 107 + 1 -UAAAGUGAGCAACCAUAAAA----ACGCGAAACAG--UUUAAAGCGUUUCGAAAUGGUAAUGAGCAAAUUACGAGAAGCCCUUCAAAGGGUUUCCACAAGCAGGUGUUUGUUU-- -..(((..((((.((......----...((((((.(--(.....))))))))...(((((((......)))))(.((((((((....))))))))).))....))))))..)))-- ( -28.60) >DroEre_CAF1 21394 106 + 1 UUAAGGUGAGCAUCCAUUAAA----ACGCGAAACAG--UUUAAAGCGUUUCGAACUGGUAAUGAGCAAAUUACGAGAAGCCCUUCAAAGGGUU--CACAAGCAGGUGUUUAUUU-- ...(((((((((((..(..((----((((.......--......))))))..).((.(((((......))))).)).((((((....))))))--........)))))))))))-- ( -27.62) >DroYak_CAF1 20706 98 + 1 CUAAAGUAA---------AAA----GCACGAAACAG--UUUAAAGCGUUUCAAAAUGGUAAUGAGCGAAUUACGAGAAGCCCUUCAAAGGGCU-CCACAAGCAGGUGUUUAUUU-- .........---------.((----(((((((((.(--(.....)))))))....(((((((......)))))..(.((((((....))))))-.).)).....))))))....-- ( -23.20) >consensus _UAAAGUGAGCAACCAUAAAA____ACGCGAAACAG__UUUAAAGCGUUUCGAAAUGGUAAUGAGCAAAUUACGAGAAGCCCUUCAAAGGGUU_CCACAAGCAGGUGUUUAUUU__ ...(((((((((.((.............((((((............)))))).....(((((......)))))....((((((....))))))..........))))))))))).. (-10.74 = -12.97 + 2.23)

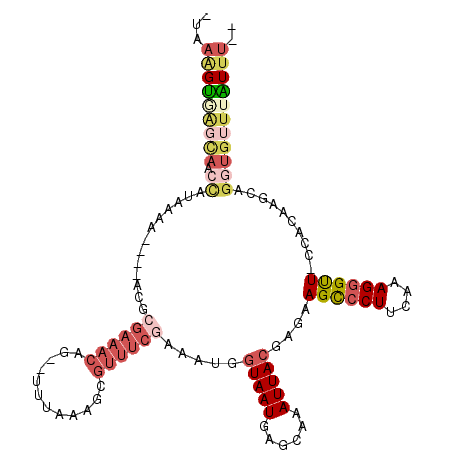

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:47 2006