
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,990,790 – 11,990,897 |
| Length | 107 |
| Max. P | 0.973163 |

| Location | 11,990,790 – 11,990,897 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
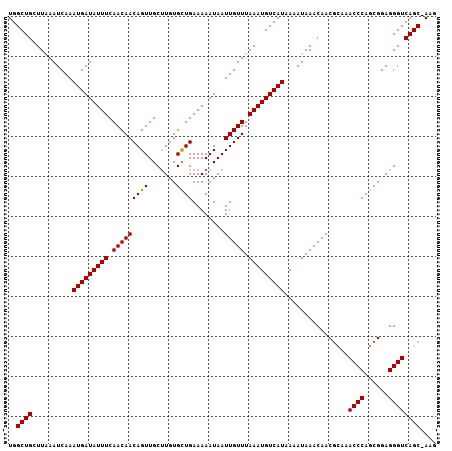
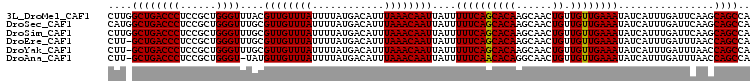
>3L_DroMel_CAF1 11990790 107 + 23771897 UGGCUGCUUGAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGUAAACCCAGCGGAGGGUCAGCCAAG ((((((..........(((((((((.(((((((((.......))))........))))).)))))))))................((((......)))))))))).. ( -26.20) >DroSec_CAF1 9197 107 + 1 UGGCUGCUUGAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGCAAACCCAGCGGAGGGUCAGCCAUG (((((((((.......(((((((((.(((((((((.......))))........))))).)))))))))...........(((.......)))..))).)))))).. ( -26.60) >DroSim_CAF1 8388 107 + 1 UGGCUGCUUGAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGCAAACCCAGCGGAGGGUCAGCCAAG (((((((((.......(((((((((.(((((((((.......))))........))))).)))))))))...........(((.......)))..))).)))))).. ( -26.80) >DroEre_CAF1 9252 106 + 1 UGGCUGGUUAAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGCAAACCCAGCGGAGGGUCAGC-AAG ..((((..(...((..(((((((((.(((((((((.......))))........))))).)))))))))...........(((.......))))).)..))))-... ( -24.40) >DroYak_CAF1 8693 106 + 1 UGGCUGGUUAAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGCAAACCCAGCGGAGGGUCAGC-AAG ..((((..(...((..(((((((((.(((((((((.......))))........))))).)))))))))...........(((.......))))).)..))))-... ( -24.40) >DroAna_CAF1 8897 105 + 1 UGGCUGGUUAAAUCAAAUGAUAUUUCAACAACAGUUGCCUGUGUUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACAUA-ACCCAGCGGAGGGUCAGC-AAG ..((((..........(((((((((.((((((((....)))((((....)))).))))).)))))))))...............-((((......))))))))-... ( -24.20) >consensus UGGCUGCUUAAAUCAAAUGAUAUUUCAACAACAGUUGCUUGUGCUGAAAAAUAAUUGUUUAAAUGUCAUAAAAUAAACAACGCAAACCCAGCGGAGGGUCAGC_AAG ..((((..........(((((((((.(((((((((.......))))........))))).)))))))))................((((......)))))))).... (-20.56 = -20.42 + -0.14)


| Location | 11,990,790 – 11,990,897 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.63 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
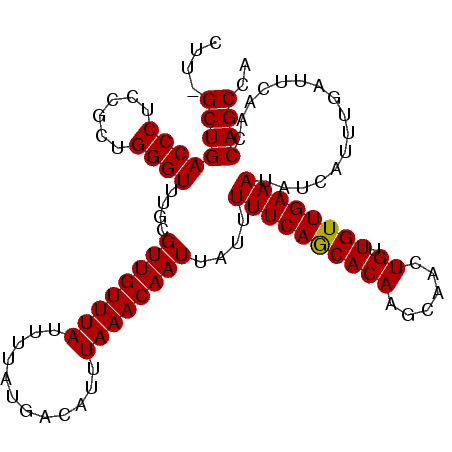
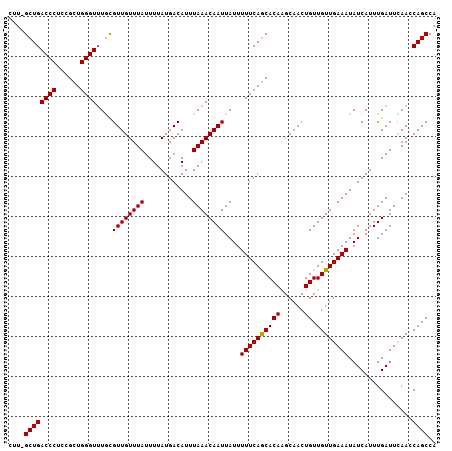
>3L_DroMel_CAF1 11990790 107 - 23771897 CUUGGCUGACCCUCCGCUGGGUUUACGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUCAAGCAGCCA ..((((((((((......))))....((((((((............))))))))....((((((((((......)).))))))))................)))))) ( -25.80) >DroSec_CAF1 9197 107 - 1 CAUGGCUGACCCUCCGCUGGGUUUGCGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUCAAGCAGCCA ..((((((((((......))))).((((((((((............))))))))....((((((((((......)).))))))))...............))))))) ( -26.90) >DroSim_CAF1 8388 107 - 1 CUUGGCUGACCCUCCGCUGGGUUUGCGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUCAAGCAGCCA ..((((((((((......))))).((((((((((............))))))))....((((((((((......)).))))))))...............))))))) ( -26.80) >DroEre_CAF1 9252 106 - 1 CUU-GCUGACCCUCCGCUGGGUUUGCGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUUAACCAGCCA ...-((.(((((......))))).))((((((((............))))))))....((((((((((......)).))))))))...................... ( -22.10) >DroYak_CAF1 8693 106 - 1 CUU-GCUGACCCUCCGCUGGGUUUGCGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUUAACCAGCCA ...-((.(((((......))))).))((((((((............))))))))....((((((((((......)).))))))))...................... ( -22.10) >DroAna_CAF1 8897 105 - 1 CUU-GCUGACCCUCCGCUGGGU-UAUGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAACACAGGCAACUGUUGUUGAAAUAUCAUUUGAUUUAACCAGCCA ...-((((((((......))))-.((((..(.......)..))))((((((((.....(((((((((((....))).))))))))......))).))))).)))).. ( -28.70) >consensus CUU_GCUGACCCUCCGCUGGGUUUGCGUUGUUUAUUUUAUGACAUUUAAACAAUUAUUUUUCAGCACAAGCAACUGUUGUUGAAAUAUCAUUUGAUUCAACCAGCCA ....((((((((......))))....((((((((............))))))))....((((((((((......)).))))))))................)))).. (-21.77 = -21.63 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:43 2006