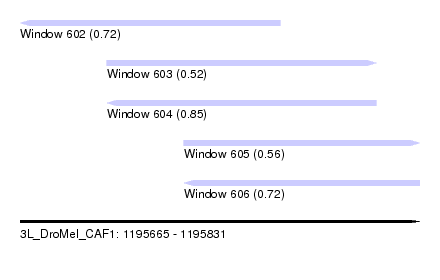
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,195,665 – 1,195,831 |
| Length | 166 |
| Max. P | 0.854354 |

| Location | 1,195,665 – 1,195,773 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.99 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1195665 108 - 23771897 UUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG--------AGCUCCAACCACCUGAAACGACUUUGUCUUGGGG----CCUCCCUUUCCU .......(((((((.......)))))))((((((((((((.((((..(((....)))((((.--------...))))............))))...))))))))----..))))...... ( -29.70) >DroSec_CAF1 12423 107 - 1 UUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG--------AGCUCCAACCACCUGAAACGACUUUGUCUUGGGG----CCUCCCUUCCC- .......(((((((.......)))))))((((((((((((.((((..(((....)))((((.--------...))))............))))...))))))))----..)))).....- ( -29.70) >DroEre_CAF1 10133 107 - 1 UUCAUUUAUGAGUUCAUCCAUAAUUCAUGGGACCCUAAGAAGGUCACGCUGGCAAGCUGGG---------AGCUCCAAACGCCUGAAUCGACUUUGUCUUAGCGG---CCUCCCUGCCC- .......(((((((.......)))))))((((((((((((.((((...(.(((....(((.---------....)))...))).)....))))...)))))).))---..)))).....- ( -27.50) >DroYak_CAF1 11942 119 - 1 UUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGGGAGCUGUUGAGCUCCAACCACCUGAAACGACUUUGUCUUAGGGGCUCCCUCCUUGCCC- .....(((((((((.......)))))))))(((.((....))))).....((((((..((((((((((((.....))))..(((((.(((....))).))))))))))))).)))))).- ( -39.70) >consensus UUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG________AGCUCCAACCACCUGAAACGACUUUGUCUUAGGG____CCUCCCUGCCC_ .......(((((((.......)))))))((((((((((((.((((..(((....)))(((((..........)))))............))))...))))))))......))))...... (-26.42 = -26.05 + -0.37)

| Location | 1,195,701 – 1,195,813 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -28.25 |
| Energy contribution | -29.12 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1195701 112 + 23771897 GGUUGGAGCU--------CUCCAGCUUGCCAGCGUGACCUUCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGCGAGUGGGGCGCUCAUCGGCGAGAUG (((((((...--------.)))))))...(((((((((((....)).)))..(((((((.......)))))))......))))))(((((((((((.......)))))))))))...... ( -38.80) >DroSec_CAF1 12458 112 + 1 GGUUGGAGCU--------CUCCAGCUUGCCAGCGUGACCUUCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGCGAGUGGAGCGCUCAUCGGCGAGAUG .((((((((.--------((((((((..((((((.(((((....)).))).)(((((((.......)))))))............)))))..)))...))))).))))..))))...... ( -39.00) >DroEre_CAF1 10169 102 + 1 GUUUGGAGCU---------CCCAGCUUGCCAGCGUGACCUUCUUAGGGUCCCAUGAAUUAUGGAUGAACUCAUAAAUGAAUGCUGGUUUGUGAGC---------GUUUGUGAGCGAGAUG .......(((---------(.(((((((((((((((((((.....))))).......((((..(((....)))..))))))))))))....))))---------...)).))))...... ( -30.80) >DroYak_CAF1 11981 111 + 1 GGUUGGAGCUCAACAGCUCCCCAGCUUGCCAGCGUGACCUUCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUUGUGAGC---------UCUCAUCGGGGGGAUG (((.((((((((..((((....)))).(((((((((((((....)).)))..(((((((.......)))))))......))))))))...)))))---------))).)))......... ( -35.20) >consensus GGUUGGAGCU________CCCCAGCUUGCCAGCGUGACCUUCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGC_________GCUCAUCGGCGAGAUG ((((((((..........)))))))).(((((((((((((....)).)))..(((((((.......)))))))......))))))))(((((((((.......)))))))))........ (-28.25 = -29.12 + 0.87)

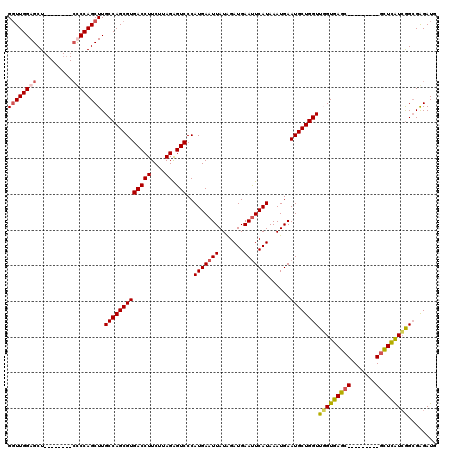
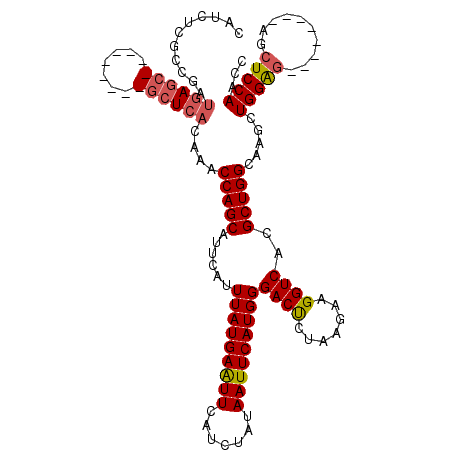
| Location | 1,195,701 – 1,195,813 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1195701 112 - 23771897 CAUCUCGCCGAUGAGCGCCCCACUCGCUCACCAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG--------AGCUCCAACC ...(((.(((.((((((.......)))))).)..(((((......(((((((((.......)))))))))(((.((....)))))..)))))......)).)--------))........ ( -30.00) >DroSec_CAF1 12458 112 - 1 CAUCUCGCCGAUGAGCGCUCCACUCGCUCACCAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG--------AGCUCCAACC ............((((.(((((...(((......(((((......(((((((((.......)))))))))(((.((....)))))..)))))..))))))))--------.))))..... ( -31.20) >DroEre_CAF1 10169 102 - 1 CAUCUCGCUCACAAAC---------GCUCACAAACCAGCAUUCAUUUAUGAGUUCAUCCAUAAUUCAUGGGACCCUAAGAAGGUCACGCUGGCAAGCUGGG---------AGCUCCAAAC ......((((.((...---------(((......(((((......(((((((((.......)))))))))((((.......))))..)))))..))))).)---------)))....... ( -25.50) >DroYak_CAF1 11981 111 - 1 CAUCCCCCCGAUGAGA---------GCUCACAAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGGGAGCUGUUGAGCUCCAACC ((((.....)))).((---------(((((....(((((......(((((((((.......)))))))))(((.((....)))))..)))))..((((....))))..)))))))..... ( -32.90) >consensus CAUCUCGCCGAUGAGC_________GCUCACAAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGAAGGUCACGCUGGCAAGCUGGAG________AGCUCCAACC ...........((((((.......))))))....(((((......(((((((((.......)))))))))((((.......))))..))))).....(((((..........)))))... (-21.70 = -22.45 + 0.75)

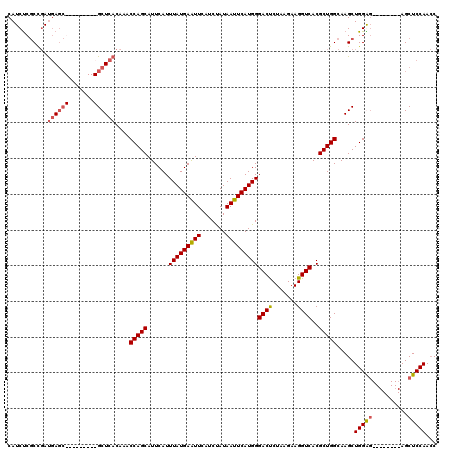
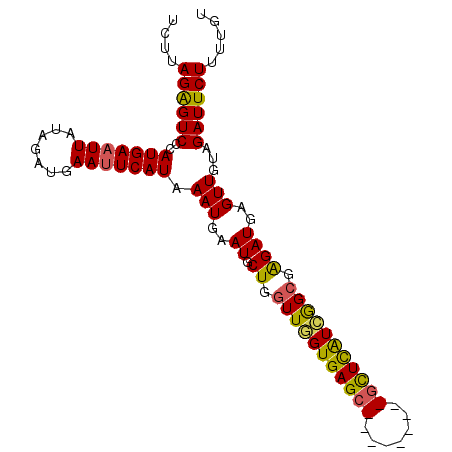
| Location | 1,195,733 – 1,195,831 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -21.02 |
| Energy contribution | -20.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1195733 98 + 23771897 UCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGCGAGUGGGGCGCUCAUCGGCGAGAUGAGUUGUAGAUUCUUUUGU ....((((((..(((((((.......))))))).(((..((.((.(((((((((((.......))))))))))).))))..)))...))))))..... ( -28.80) >DroSec_CAF1 12490 98 + 1 UCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGCGAGUGGAGCGCUCAUCGGCGAGAUGAGUUGUAGAUUCUUUUGU ....((((((..(((((((.......))))))).(((..((.((.(((((((((((.......))))))))))).))))..)))...))))))..... ( -28.80) >DroEre_CAF1 10200 89 + 1 UCUUAGGGUCCCAUGAAUUAUGGAUGAACUCAUAAAUGAAUGCUGGUUUGUGAGC---------GUUUGUGAGCGAGAUGAGUUGUCGAUUCUUUUGU ....(((((((((((...)))))...(((((((..(..((((((.(....).)))---------)))..).......)))))))...))))))..... ( -21.10) >DroYak_CAF1 12021 89 + 1 UCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUUGUGAGC---------UCUCAUCGGGGGGAUGAGUUGUCGAUUCUUUUGU ....((((((.((..(((..(((....(((((....))))).))))))..)).((---------.((((((.....))))))..)).))))))..... ( -19.50) >consensus UCUUAGAGUCCCAUGAAUUAUAGAUGAAUUCAUAAAUGAAUGCUGGUUGGUGAGC_________GCUCAUCGGCGAGAUGAGUUGUAGAUUCUUUUGU ....((((((..(((((((.......))))))).(((..((.((.(((((((((((.......))))))))))).))))..)))...))))))..... (-21.02 = -20.78 + -0.25)

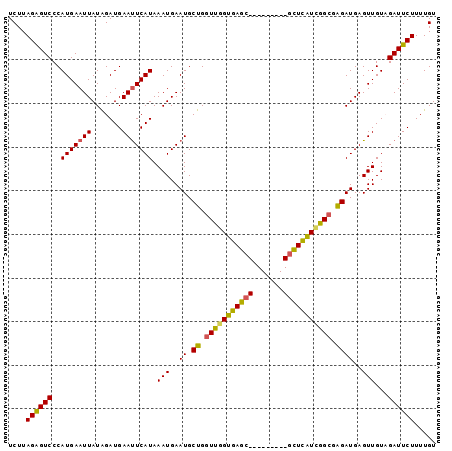
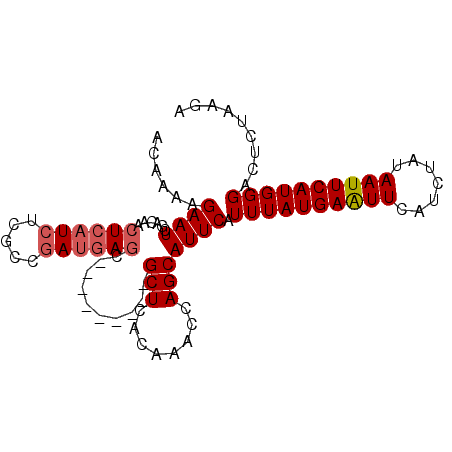
| Location | 1,195,733 – 1,195,831 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -14.82 |
| Consensus MFE | -11.46 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1195733 98 - 23771897 ACAAAAGAAUCUACAACUCAUCUCGCCGAUGAGCGCCCCACUCGCUCACCAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGA .....(((........((((((.....))))))...(((....(((........)))........(((((((.......))))))))))..))).... ( -17.70) >DroSec_CAF1 12490 98 - 1 ACAAAAGAAUCUACAACUCAUCUCGCCGAUGAGCGCUCCACUCGCUCACCAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGA .....(((........((((((.....))))))...(((....(((........))).......((((((((.......))))))))))).))).... ( -16.60) >DroEre_CAF1 10200 89 - 1 ACAAAAGAAUCGACAACUCAUCUCGCUCACAAAC---------GCUCACAAACCAGCAUUCAUUUAUGAGUUCAUCCAUAAUUCAUGGGACCCUAAGA ..............(((((((...(....)....---------(((........)))........)))))))..(((((.....)))))......... ( -8.70) >DroYak_CAF1 12021 89 - 1 ACAAAAGAAUCGACAACUCAUCCCCCCGAUGAGA---------GCUCACAAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGA .....(((.((((...((((((.....)))))).---------(((........)))..))..(((((((((.......))))))))))).))).... ( -16.30) >consensus ACAAAAGAAUCGACAACUCAUCUCGCCGAUGAGC_________GCUCACAAACCAGCAUUCAUUUAUGAAUUCAUCUAUAAUUCAUGGGACUCUAAGA ......((((......((((((.....))))))..........(((........))))))).((((((((((.......))))))))))......... (-11.46 = -12.28 + 0.81)

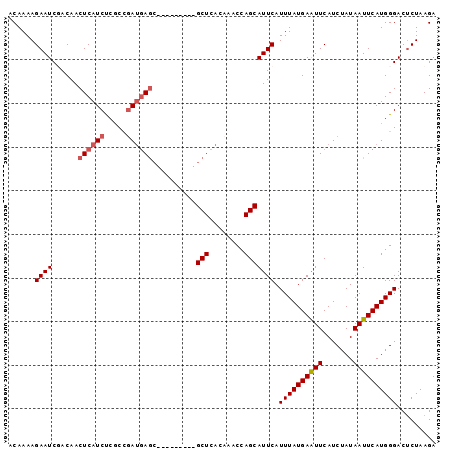
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:16 2006