
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,988,667 – 11,988,778 |
| Length | 111 |
| Max. P | 0.997015 |

| Location | 11,988,667 – 11,988,778 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -36.98 |
| Energy contribution | -37.86 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
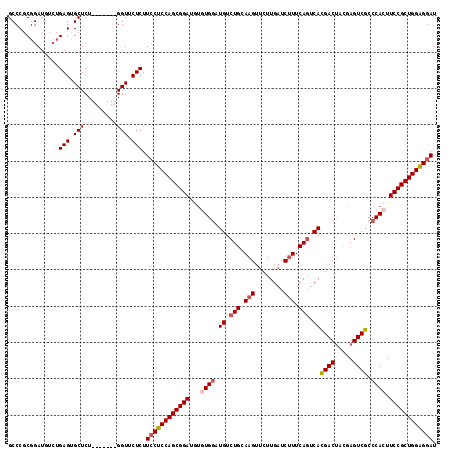
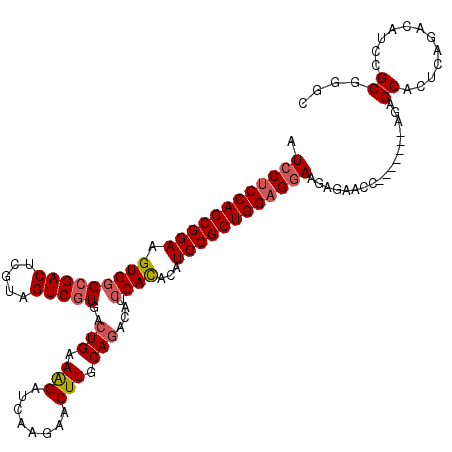
>3L_DroMel_CAF1 11988667 111 + 23771897 GCCCGCGGAUGUCUGAGUGCUCU-------GGUUCUCUUCCUCCAGCGGAUAUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGGGGAU ((.(.(((....))).).))...-------........((((((((((((...((((.((.(((.(((........))).))).))((((.....)))).)))).)))))))))))). ( -44.30) >DroSec_CAF1 7047 111 + 1 GCCCGCGGAUGUCUGAGUGCUCU-------GGUUCUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGAGGAU ((.(.(((....))).).))...-------........((((((((((((.(((.((.((.(((.(((........))).))).))((((.....))))))))).)))))))))))). ( -45.50) >DroSim_CAF1 6246 111 + 1 GCCCGCGGAUGUCUGAGUGCUCU-------GGUUCUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACCACGAGUCGCCCACUUCCGCUGGAGGAU ((.(.(((....))).).))...-------........((((((((((((.(((.((.((.(((.(((........))).))).))((((.....))))))))).)))))))))))). ( -44.80) >DroEre_CAF1 7104 107 + 1 GCCCGCGGAU----GAGCGCUCU-------GGUUCUCUUGCUCCAGCGGAUACAUGGCUGUCUGGAAGCUCUUGUUCCUUCAGUCAUGACUAUGAGUCGCCCACUUCCGCUGGAGGAU ..((((((..----((((.....-------.))))..))))(((((((((...(((((((...((((.(....)))))..)))))))(((.....))).......))))))))))).. ( -41.80) >DroYak_CAF1 6376 118 + 1 GCCCGCGGAUGUCUGAGUGCUCUGGUUUCUGGUUCUCUUCCUCCAGCGGAUGUAUGCAUGUCUGGAAGUUCUUGAUCUUUCACUCACGACUAUGAGUCGCCCACUUCCGCUGGAGGAU .....((((..((.((....)).))..)))).......((((((((((((.((..((.((..((((((........))))))..)).(((.....)))))..)).)))))))))))). ( -42.80) >consensus GCCCGCGGAUGUCUGAGUGCUCU_______GGUUCUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGAGGAU ..............(((.(((.........))).))).((((((((((((...((((.((.(((.(((........))).))).))((((.....)))).)))).)))))))))))). (-36.98 = -37.86 + 0.88)


| Location | 11,988,667 – 11,988,778 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -39.26 |
| Consensus MFE | -34.54 |
| Energy contribution | -35.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11988667 111 - 23771897 AUCCCCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACAUAUCCGCUGGAGGAAGAGAACC-------AGAGCACUCAGACAUCCGCGGGC .(((.((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...)))))))).)))......((-------...((............)).)). ( -37.30) >DroSec_CAF1 7047 111 - 1 AUCCUCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACACAUCCGCUGGAGGAAGAGAACC-------AGAGCACUCAGACAUCCGCGGGC .((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...))))))))))))......((-------...((............)).)). ( -42.10) >DroSim_CAF1 6246 111 - 1 AUCCUCCAGCGGAAGUGGGCGACUCGUGGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACACAUCCGCUGGAGGAAGAGAACC-------AGAGCACUCAGACAUCCGCGGGC .((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...))))))))))))......((-------...((............)).)). ( -42.20) >DroEre_CAF1 7104 107 - 1 AUCCUCCAGCGGAAGUGGGCGACUCAUAGUCAUGACUGAAGGAACAAGAGCUUCCAGACAGCCAUGUAUCCGCUGGAGCAAGAGAACC-------AGAGCGCUC----AUCCGCGGGC ...((((((((((....((((((.....))).((.(((.(((........))).))).))))).....))))))))))........((-------...(((...----...))).)). ( -36.60) >DroYak_CAF1 6376 118 - 1 AUCCUCCAGCGGAAGUGGGCGACUCAUAGUCGUGAGUGAAAGAUCAAGAACUUCCAGACAUGCAUACAUCCGCUGGAGGAAGAGAACCAGAAACCAGAGCACUCAGACAUCCGCGGGC .((((((((((((.(((.(((((((((....))))))(.(((........))).).....))).))).)))))))))))).............((...((............)).)). ( -38.10) >consensus AUCCUCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACACAUCCGCUGGAGGAAGAGAACC_______AGAGCACUCAGACAUCCGCGGGC .((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...))))))))))))..................((............)).... (-34.54 = -35.14 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:41 2006