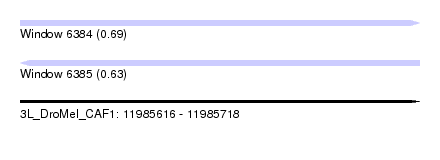
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,985,616 – 11,985,718 |
| Length | 102 |
| Max. P | 0.690953 |

| Location | 11,985,616 – 11,985,718 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -41.69 |
| Consensus MFE | -25.11 |
| Energy contribution | -26.53 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11985616 102 + 23771897 ACAUAUGGUGGAUCCGGUUCCUGCAGCGGCACCUCAAAGAACAGUGGCUUCAGGGCCAACUGCACAGCCGCCAAACUGGAGGCUGUCA---CCCUGGAUC---CCACU ......((((((((((((.((((.(((.((.............)).))).)))))))...((.((((((.((.....)).))))))))---....)))).---))))) ( -41.02) >DroSim_CAF1 3173 102 + 1 ACAUAUGGUGGAUCCGGUUCCUGCAGCGGCACCUCGAAGAACAGUGGCUUCAGGGCCAACUGCACAGCCGCCAAACUGGAGGCUGUCA---CCCGGGAUC---CCACU ......(((((((((((.....((((.(((.(((.((((........))))))))))..))))((((((.((.....)).))))))..---.)).)))).---))))) ( -43.60) >DroEre_CAF1 4002 102 + 1 ACGUAUGGAGGAUCCGGCUCCUGCAGCGGCACCUCAAAAAACAAUGGCUUCAGGGCCAGCUGAACAGCCGCCAGGCUGGAGGCUGUCA---CCCUGGAUC---CCACU ......(..((((((((((.....)))((((((((......((.(((((....)))))..))..(((((....))))))))).)))).---...))))))---)..). ( -41.50) >DroWil_CAF1 4424 99 + 1 ACAUACGGUGGGUCGGGUUCCUGCAGCGGCACCUCAAAGAACAAGGGUUUCAGAGCCAGCUGAACGGCUGCGUGAGCAGCUGCUGCCG---CUCUG---C---CCCCA ......((.(((.(((((....))((((((((((.........)))..(((((......)))))(((((((....))))))).)))))---)))))---)---)))). ( -42.00) >DroYak_CAF1 3174 102 + 1 ACGUACGGAGGAUCUGGCUCCUGCAGCGGCACCUCAAAGAACAACGGCUUCAGGGCCAACUGAACAGCCGCCAAGGUGGAGGCUGUCA---CCCUGGAAC---CCACU ..((.(.((((..(((((....))..)))..))))...).))...((.(((((((.....(((.(((((.((.....)).))))))))---))))))).)---).... ( -34.80) >DroAna_CAF1 3988 108 + 1 AAGUACGGAGGAUCCGGCUCCUGCAGGGGCACCUCAAAGAAGAGUGGCUUGAGGGCCAACUGGACAGCCGCCAGAGUAGAGGCUGUAAGUCCCCUGGAUCCUCCCGCU .(((..(((((((((.((....))(((((...(((......)))(((((....)))))(((..((((((...........)))))).))))))))))))))))).))) ( -47.20) >consensus ACAUACGGAGGAUCCGGCUCCUGCAGCGGCACCUCAAAGAACAAUGGCUUCAGGGCCAACUGAACAGCCGCCAAACUGGAGGCUGUCA___CCCUGGAUC___CCACU ......((..((((((((.((((.(((.(...............).))).)))))))......((((((.((.....)).)))))).........)))))...))... (-25.11 = -26.53 + 1.42)

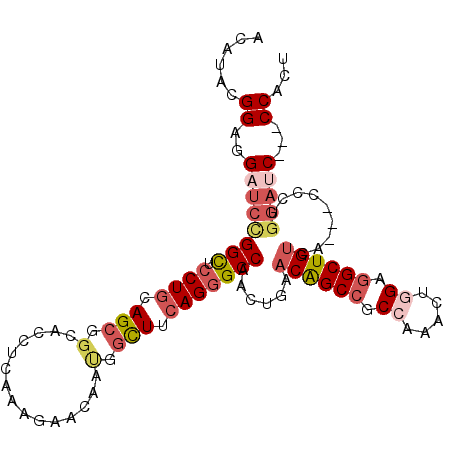
| Location | 11,985,616 – 11,985,718 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -28.25 |
| Energy contribution | -29.53 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
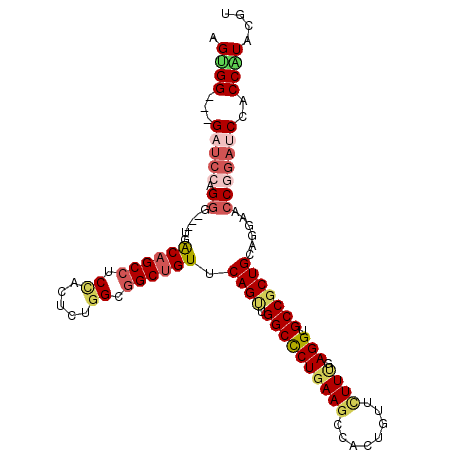
>3L_DroMel_CAF1 11985616 102 - 23771897 AGUGG---GAUCCAGGG---UGACAGCCUCCAGUUUGGCGGCUGUGCAGUUGGCCCUGAAGCCACUGUUCUUUGAGGUGCCGCUGCAGGAACCGGAUCCACCAUAUGU ....(---(((((.((.---..((((((.((.....)).))))))(((((.((((((((((........)))).))).)))))))).....))))))))......... ( -43.90) >DroSim_CAF1 3173 102 - 1 AGUGG---GAUCCCGGG---UGACAGCCUCCAGUUUGGCGGCUGUGCAGUUGGCCCUGAAGCCACUGUUCUUCGAGGUGCCGCUGCAGGAACCGGAUCCACCAUAUGU ....(---(((((.((.---..((((((.((.....)).))))))(((((.((((((((((........)))).))).)))))))).....))))))))......... ( -46.10) >DroEre_CAF1 4002 102 - 1 AGUGG---GAUCCAGGG---UGACAGCCUCCAGCCUGGCGGCUGUUCAGCUGGCCCUGAAGCCAUUGUUUUUUGAGGUGCCGCUGCAGGAGCCGGAUCCUCCAUACGU .(..(---(((((.((.---((((((((.((.....)).))))).))).))(((((((.(((((((.........))))..))).)))).)))))))))..)...... ( -45.70) >DroWil_CAF1 4424 99 - 1 UGGGG---G---CAGAG---CGGCAGCAGCUGCUCACGCAGCCGUUCAGCUGGCUCUGAAACCCUUGUUCUUUGAGGUGCCGCUGCAGGAACCCGACCCACCGUAUGU .((((---(---(((((---(((((((.(((((....))))).)))..))).)))))).............(((.(((.((......)).))))))))).))...... ( -37.00) >DroYak_CAF1 3174 102 - 1 AGUGG---GUUCCAGGG---UGACAGCCUCCACCUUGGCGGCUGUUCAGUUGGCCCUGAAGCCGUUGUUCUUUGAGGUGCCGCUGCAGGAGCCAGAUCCUCCGUACGU .((((---(....((((---((((((((.((.....)).)))))))...(((((((((.(((.((..(((...)))..)).))).)))).)))))))))))).))).. ( -39.10) >DroAna_CAF1 3988 108 - 1 AGCGGGAGGAUCCAGGGGACUUACAGCCUCUACUCUGGCGGCUGUCCAGUUGGCCCUCAAGCCACUCUUCUUUGAGGUGCCCCUGCAGGAGCCGGAUCCUCCGUACUU ....((((((((((((((........))))).....(((..(((..(((..((((((((((.........))))))).))).))))))..))))))))))))...... ( -47.80) >consensus AGUGG___GAUCCAGGG___UGACAGCCUCCACUCUGGCGGCUGUUCAGUUGGCCCUGAAGCCACUGUUCUUUGAGGUGCCGCUGCAGGAACCGGAUCCACCAUACGU .((((...(((((.((......((((((.((.....)).)))))).((((.((((((((((........)))).))).)))))))......)))))))..)))).... (-28.25 = -29.53 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:35 2006