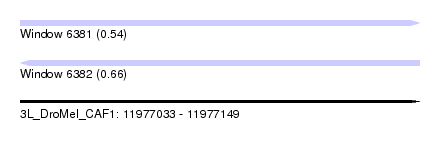
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,977,033 – 11,977,149 |
| Length | 116 |
| Max. P | 0.656387 |

| Location | 11,977,033 – 11,977,149 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -23.09 |
| Energy contribution | -25.90 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
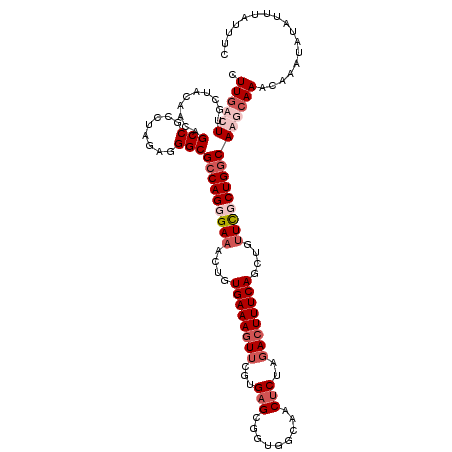
>3L_DroMel_CAF1 11977033 116 + 23771897 UUUGACUCGCUAUAGCAGCCGCCUAGAGGGCGCCAGGGAAACUAUGAAAGUUGGUGAGCGAUGGCAACACUAGACUUUCAGCUGUUCGCUGACAGUCAAACAAAUAUAUUUAUUUC ((((..(((((.((.((((((((.....))))...((....))......)))).)))))))((....))...((((.(((((.....))))).))))...))))............ ( -35.00) >DroSim_CAF1 7307 116 + 1 AUUGACUUGCAAUAGCAGCCGCCUAGAGGGCGCCAGGGAAACUGUGAAAAUUCGUGAGCGGUGACAACUCUAGACUUUCAGCUGUUCGCUGGCAGUCAAACAAAUAUAUUUAUUUC .(((((((((....)))...(((.....)))(((((.(((...((..(((.((..(((.(....)..)))..)).)))..))..))).)))))))))))................. ( -33.10) >DroEre_CAF1 4944 115 + 1 CUUGACUUGCUACAACUGCCGCCUAGAGGGCGCCAGUGA-ACUGUGAAAGUUCGUGAGCGGUGGCAGCUCUGUACUUUCAGCUGUUUGCUGGCAAGCAAGCAAAUUUAUUUAUUUC .(((.((((((.........(((.....)))(((((..(-((.((((((((.((.((((.......)))))).))))))).).)))..))))).)))))))))............. ( -45.70) >DroYak_CAF1 4720 114 + 1 CUU-ACUUGCUACAACCGCCGCC-AGAGGGCGCCAGUGAAACUGUGAAAGUUCGUGAGCGGUGGCAACUCUUGACUUUCAGCUGUUCGCUGGCAAGCAAACAAAUUUAUUUAUUUC ...-..(((((.........(((-....)))(((((((((....((((((((...(((..(...)..)))..))))))))....))))))))).)))))................. ( -35.90) >consensus CUUGACUUGCUACAACAGCCGCCUAGAGGGCGCCAGGGAAACUGUGAAAGUUCGUGAGCGGUGGCAACUCUAGACUUUCAGCUGUUCGCUGGCAAGCAAACAAAUAUAUUUAUUUC .((((((..........(((........)))(((((((((....((((((((...(((.........)))..))))))))....)))))))))))))))................. (-23.09 = -25.90 + 2.81)

| Location | 11,977,033 – 11,977,149 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -25.34 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
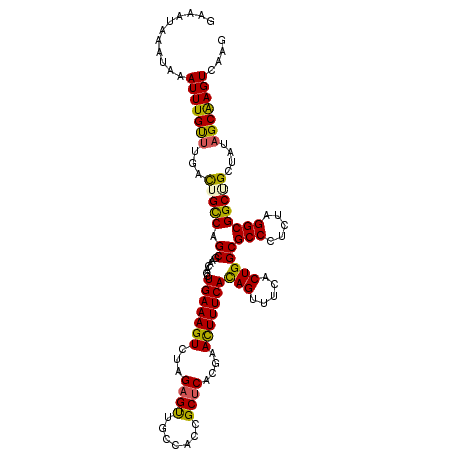
>3L_DroMel_CAF1 11977033 116 - 23771897 GAAAUAAAUAUAUUUGUUUGACUGUCAGCGAACAGCUGAAAGUCUAGUGUUGCCAUCGCUCACCAACUUUCAUAGUUUCCCUGGCGCCCUCUAGGCGGCUGCUAUAGCGAGUCAAA ...........(((((((.((((.(((((.....))))).))))((((...(((...(((....((((.....)))).....)))(((.....)))))).)))).))))))).... ( -30.80) >DroSim_CAF1 7307 116 - 1 GAAAUAAAUAUAUUUGUUUGACUGCCAGCGAACAGCUGAAAGUCUAGAGUUGUCACCGCUCACGAAUUUUCACAGUUUCCCUGGCGCCCUCUAGGCGGCUGCUAUUGCAAGUCAAU .................((((((((((((.....)))....((((((((.......((((...(((..........)))...))))..)))))))))))(((....))))))))). ( -30.20) >DroEre_CAF1 4944 115 - 1 GAAAUAAAUAAAUUUGCUUGCUUGCCAGCAAACAGCUGAAAGUACAGAGCUGCCACCGCUCACGAACUUUCACAGU-UCACUGGCGCCCUCUAGGCGGCAGUUGUAGCAAGUCAAG ................(((((((((((((.....))))....((((..((((((...(((...(((((.....)))-))...)))(((.....)))))))))))))))))).)))) ( -38.80) >DroYak_CAF1 4720 114 - 1 GAAAUAAAUAAAUUUGUUUGCUUGCCAGCGAACAGCUGAAAGUCAAGAGUUGCCACCGCUCACGAACUUUCACAGUUUCACUGGCGCCCUCU-GGCGGCGGUUGUAGCAAGU-AAG .................((((((((((((.....))))............(((.(((((.............(((.....))).((((....-))))))))).)))))))))-)). ( -34.30) >consensus GAAAUAAAUAAAUUUGUUUGACUGCCAGCGAACAGCUGAAAGUCUAGAGUUGCCACCGCUCACGAACUUUCACAGUUUCACUGGCGCCCUCUAGGCGGCUGCUAUAGCAAGUCAAG ...........(((((((...(((((.((.......(((((((...((((.......))))....)))))))(((.....)))))(((.....))))))))....))))))).... (-25.34 = -24.90 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:32 2006