
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,952,637 – 11,952,743 |
| Length | 106 |
| Max. P | 0.960036 |

| Location | 11,952,637 – 11,952,743 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11952637 106 + 23771897 CG--UAACGCGUUUCCAAUAACUAUCCUUUUAUUUCGCUAU---UACAGUCCGGAAACUGCGAUCCUCAGGUCUGCAAUGACCAGUCAGAGUACUUCGUCUCUCCGGACUA .(--(((.(((.....(((((........))))).)))..)---)))((((((((....((((..(((.((((......)))).....)))....))))...)))))))). ( -27.50) >DroSec_CAF1 5514 106 + 1 CG--UAACGCCUUUCCAAUAACUAUCCCUUUAUUUCUCUAU---UACAGUCCGGAAACUGCGAUCCUCUGGUCUGCAAUGACCAGUCGGAGUACUUCGUCUCUCCGGACUA ..--.....................................---...((((((((....(((((((.((((((......))))))..))).....))))...)))))))). ( -29.50) >DroSim_CAF1 5452 106 + 1 CG--UAACGCCUUUCCAAUAACUAUCCCUUUAUUUCGCUAU---UACAGUCCGGAAACUGCGAUCCUCUGGUCUGCAAUGACCAGUCGGAGUACUUCGUCUCUCCGGACUA .(--(((.((......(((((........)))))..))..)---)))((((((((....(((((((.((((((......))))))..))).....))))...)))))))). ( -29.80) >DroEre_CAF1 5973 107 + 1 CGACCUACAUGUUUUCAACAACAAUCCCUUCAU-UCCCUAU---UACAGUCCGGAAACUGCGAUCCCCGGGUCUGCACCGGCAAGUUGGAUUUCUACGUCUCUCCGGACUU .........((((......))))..........-.......---...((((((((...(((((((....)))).)))((((....)))).............)))))))). ( -23.80) >DroYak_CAF1 3652 108 + 1 CGACCUACAAGUUUUCAACAACAAUCCCCUUAUUUCCGUAU---UACAGUCCGGAAACUGCGAUCCUCAGGUCUGCGUCGACCAGACGGAGUACUACAUCUCUCCGGACUA .........................................---...((((((((..(((.......)))((((.((((.....)))).)).))........)))))))). ( -23.70) >DroPer_CAF1 6244 105 + 1 ------AGAAAUUUUGGAUGAGUAACAUUAUAUCCCGCUUUAAACGCAGACUGGAGAGUGCGAUCCCGCGUCCUGUGUCGGUAAUCCGGAGUACUGGAUUUCCCCCAAUUA ------.......((((..........................(((((((((((.((......))))).)).)))))).((.(((((((....))))))).)).))))... ( -25.00) >consensus CG__UAACACGUUUCCAAUAACUAUCCCUUUAUUUCGCUAU___UACAGUCCGGAAACUGCGAUCCUCAGGUCUGCAACGACCAGUCGGAGUACUUCGUCUCUCCGGACUA ...............................................(((((((....(((((((....)))).)))..........((((........))))))))))). (-16.30 = -16.42 + 0.12)

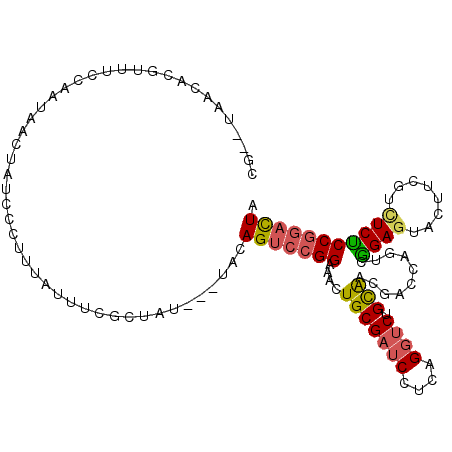
| Location | 11,952,637 – 11,952,743 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11952637 106 - 23771897 UAGUCCGGAGAGACGAAGUACUCUGACUGGUCAUUGCAGACCUGAGGAUCGCAGUUUCCGGACUGUA---AUAGCGAAAUAAAAGGAUAGUUAUUGGAAACGCGUUA--CG .((((((((((..(((....(((.....((((......)))).)))..)))...))))))))))(((---((.((..(((((........)))))(....)))))))--). ( -32.30) >DroSec_CAF1 5514 106 - 1 UAGUCCGGAGAGACGAAGUACUCCGACUGGUCAUUGCAGACCAGAGGAUCGCAGUUUCCGGACUGUA---AUAGAGAAAUAAAGGGAUAGUUAUUGGAAAGGCGUUA--CG ..((((((((..........))))..((((((......)))))).))))(((..(((((.((((((.---....(....)......))))))...))))).)))...--.. ( -31.80) >DroSim_CAF1 5452 106 - 1 UAGUCCGGAGAGACGAAGUACUCCGACUGGUCAUUGCAGACCAGAGGAUCGCAGUUUCCGGACUGUA---AUAGCGAAAUAAAGGGAUAGUUAUUGGAAAGGCGUUA--CG (((((((((((..(((.....(((..((((((......)))))).))))))...)))))))))))((---(((((..............)))))))...........--.. ( -33.84) >DroEre_CAF1 5973 107 - 1 AAGUCCGGAGAGACGUAGAAAUCCAACUUGCCGGUGCAGACCCGGGGAUCGCAGUUUCCGGACUGUA---AUAGGGA-AUGAAGGGAUUGUUGUUGAAAACAUGUAGGUCG ...........(((.((.((((((..(((.((..(((((..((((..((....))..)))).)))))---...)).)-).)...)))))(((......))).).)).))). ( -29.70) >DroYak_CAF1 3652 108 - 1 UAGUCCGGAGAGAUGUAGUACUCCGUCUGGUCGACGCAGACCUGAGGAUCGCAGUUUCCGGACUGUA---AUACGGAAAUAAGGGGAUUGUUGUUGAAAACUUGUAGGUCG (((((((((((..((..((.(((.((((((....).)))))..))).))..)).)))))))))))..---...(((..(((((.................)))))...))) ( -32.13) >DroPer_CAF1 6244 105 - 1 UAAUUGGGGGAAAUCCAGUACUCCGGAUUACCGACACAGGACGCGGGAUCGCACUCUCCAGUCUGCGUUUAAAGCGGGAUAUAAUGUUACUCAUCCAAAAUUUCU------ ...(((((((.(((((........))))).))((((..(((.(((....)))....)))..(((((.......)))))......)))).....))))).......------ ( -23.90) >consensus UAGUCCGGAGAGACGAAGUACUCCGACUGGUCAUUGCAGACCAGAGGAUCGCAGUUUCCGGACUGUA___AUAGCGAAAUAAAGGGAUAGUUAUUGAAAACGCGUUA__CG (((((((((((......((..(((....((((......))))...)))..))..))))))))))).............................................. (-18.54 = -19.27 + 0.73)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:25 2006