
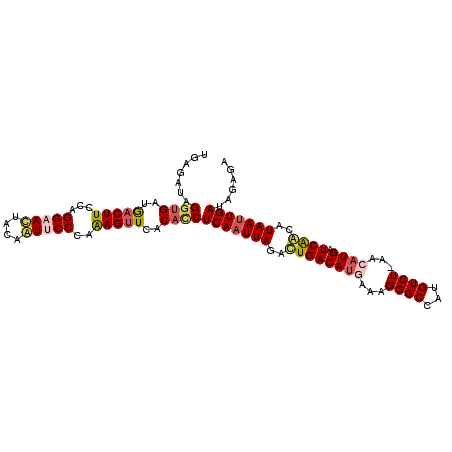
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,949,890 – 11,949,995 |
| Length | 105 |
| Max. P | 0.996963 |

| Location | 11,949,890 – 11,949,995 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -22.73 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11949890 105 + 23771897 UGAAAUAGGUGAUGACUUCCAGGAAUUACAAUUCCCAAAGUUCACACCUCAAUUAGACUGACAUGAAAGGGCCAUGUCU-AACAUGAUCGGCAUAAUUGAUAGAGA .......((((.((((((...(((((....)))))..))).)))))))(((((((..((((((((...((((...))))-..)))).))))..)))))))...... ( -28.00) >DroSec_CAF1 2766 105 + 1 UGAGAUAGGUGAUGACUUCCAGGAAUUACGAUUCCCAAAGUUCACACCUCAAUUAGACUGACAUAAAAGGGCCAUGUCU-AACAUGAUCAGCAUAAAUGAUAGAGA .......((((.((((((...(((((....)))))..))).)))))))(((.(((..(((((((....((((...))))-...))).))))..))).)))...... ( -22.00) >DroSim_CAF1 2771 105 + 1 UGAGAUAGGUGAUGACUUCCAGGAAUAACGAUUCCCAAAGUUCACACCUCAAUUAGACUGACAUAUAAGGGCCAUGUCU-AACAUGAUCAGCAUAAUUGAUAGAGA .......((((.((((((...(((((....)))))..))).)))))))(((((((..(((((((....((((...))))-...))).))))..)))))))...... ( -25.80) >DroEre_CAF1 3168 105 + 1 UGCGAUAGGUGAUGACUUCUAGGAACUACAAUGCCCAAAGUUCACAUCUCAAUUAGACUGACACGAAAGGGCCAUGUCU-AACAUGAUCAACGUAAUUGACAGAGA ((((...(((.(((........(((((...........))))).........((((((((.(.(....).).)).))))-))))).)))..))))........... ( -19.00) >DroYak_CAF1 919 105 + 1 UGCGAUAGGUGAUGACUUCCAGGAACUACAGUGCCCAAAGUUCACACCUCAAUUAGAUUGACAUGAAAGGGCCAUGUCU-AACAUGAUCAACGUAAUUGAUAGAGA .......((((.((((((...((.((....)).))..))).)))))))(((((((..((((((((...((((...))))-..)))).))))..)))))))...... ( -24.40) >DroAna_CAF1 2773 101 + 1 U-----AGAAGUCAACUUCUAGGAACCCAGGUUCCAAGAGAUCACACCUCAAUUAGGUUGACAUGAUAGGGCUGUGUCUUGACAUGAUCAACAUAAUUGAUGGCGA (-----(((((....))))))(((((....))))).........(.(((((((((.(((((((((.((((((...)))))).)))).))))).))))))).)).). ( -36.80) >consensus UGAGAUAGGUGAUGACUUCCAGGAACUACAAUUCCCAAAGUUCACACCUCAAUUAGACUGACAUGAAAGGGCCAUGUCU_AACAUGAUCAACAUAAUUGAUAGAGA .......((((..(((((...(((((....)))))..)))))..))))(((((((..((((((((...((((...))))...)))).))))..)))))))...... (-22.73 = -23.37 + 0.64)


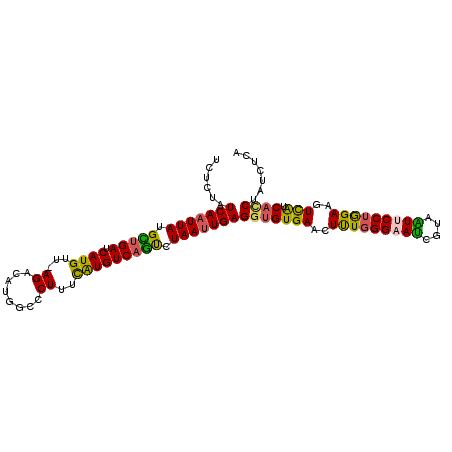
| Location | 11,949,890 – 11,949,995 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -29.04 |
| Energy contribution | -28.02 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11949890 105 - 23771897 UCUCUAUCAAUUAUGCCGAUCAUGUU-AGACAUGGCCCUUUCAUGUCAGUCUAAUUGAGGUGUGAACUUUGGGAAUUGUAAUUCCUGGAAGUCAUCACCUAUUUCA ......(((((((.((.((.((((..-((........))..)))))).)).)))))))(((((((..((..(((((....)))))..))..))).))))....... ( -31.20) >DroSec_CAF1 2766 105 - 1 UCUCUAUCAUUUAUGCUGAUCAUGUU-AGACAUGGCCCUUUUAUGUCAGUCUAAUUGAGGUGUGAACUUUGGGAAUCGUAAUUCCUGGAAGUCAUCACCUAUCUCA ......(((.(((.(((((.((((..-((........))..))))))))).))).)))(((((((..((..(((((....)))))..))..))).))))....... ( -30.90) >DroSim_CAF1 2771 105 - 1 UCUCUAUCAAUUAUGCUGAUCAUGUU-AGACAUGGCCCUUAUAUGUCAGUCUAAUUGAGGUGUGAACUUUGGGAAUCGUUAUUCCUGGAAGUCAUCACCUAUCUCA ......(((((((.(((((.(((((.-((........)).)))))))))).)))))))(((((((..((..(((((....)))))..))..))).))))....... ( -34.30) >DroEre_CAF1 3168 105 - 1 UCUCUGUCAAUUACGUUGAUCAUGUU-AGACAUGGCCCUUUCGUGUCAGUCUAAUUGAGAUGUGAACUUUGGGCAUUGUAGUUCCUAGAAGUCAUCACCUAUCGCA ......(((((((..((((.((((..-((........))..))))))))..)))))))(((((((((((((((.((....)).))))).)))..))))..)))... ( -23.40) >DroYak_CAF1 919 105 - 1 UCUCUAUCAAUUACGUUGAUCAUGUU-AGACAUGGCCCUUUCAUGUCAAUCUAAUUGAGGUGUGAACUUUGGGCACUGUAGUUCCUGGAAGUCAUCACCUAUCGCA ......(((((((.(((((.((((..-((........))..))))))))).)))))))(((((((..((..((.((....)).))..))..))).))))....... ( -31.60) >DroAna_CAF1 2773 101 - 1 UCGCCAUCAAUUAUGUUGAUCAUGUCAAGACACAGCCCUAUCAUGUCAACCUAAUUGAGGUGUGAUCUCUUGGAACCUGGGUUCCUAGAAGUUGACUUCU-----A .((((.(((((((.(((((.((((...((........))..))))))))).))))))))))).........(((((....)))))((((((....)))))-----) ( -32.80) >consensus UCUCUAUCAAUUAUGCUGAUCAUGUU_AGACAUGGCCCUUUCAUGUCAGUCUAAUUGAGGUGUGAACUUUGGGAAUCGUAAUUCCUGGAAGUCAUCACCUAUCUCA ......(((((((.(((((.((((...((........))..))))))))).)))))))(((((((..(((((((((....)))))))))..))).))))....... (-29.04 = -28.02 + -1.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:22 2006