
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,920,048 – 11,920,151 |
| Length | 103 |
| Max. P | 0.981333 |

| Location | 11,920,048 – 11,920,151 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
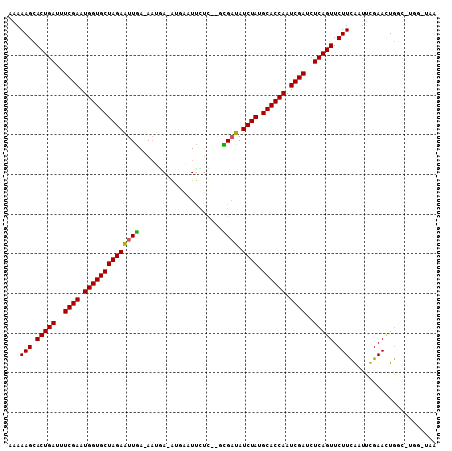
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11920048 103 + 23771897 CAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUAA-AUUGA-GUGGGUUCUC--CUGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAAGUGGC-AGGGCAA .....(((((((..((((.((((((((((.....-...((-(......)))--......)))).)))))).))))..))))).(((((...)))))..))-....... ( -32.34) >DroVir_CAF1 23888 104 + 1 AAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGA-AAUGA-AUUACUUUUUGGGCGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGC-UGG-UGC .....(((((....((((.((((((((((((((.-..(((-(......))))..)))).)))).)))))).))))..(((((((........))))))).-.))-))) ( -34.20) >DroPse_CAF1 2703 104 + 1 CAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAAAUUA-GCGAGUCCUC--UUGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAACUGUA-AUUGUAA ...(((.(((((..((((.(((((((((((((((...)))-))(((....)--))....)))).)))))).))))..))))).)))..............-....... ( -28.80) >DroGri_CAF1 12732 104 + 1 ACAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGA-AAUUAAAUU-AUUCUUGGACGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGC-UGC-UAC ....((((((((..((((.((((((((((((((.-.........-.........)))).)))).)))))).))))..))(((((........))))))).-)))-).. ( -29.11) >DroMoj_CAF1 14665 106 + 1 AAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGA-AAUAAAAUGACUUUUUGGGCGUUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGUGUGG-UGU .....(((((....((((.((((((((((..((.-..(((((.....)))))..))...)))).)))))).))))..(((((((........)))))))...))-))) ( -32.10) >DroAna_CAF1 2225 103 + 1 CAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGA-AAUGA-UCGGAUCUCC--UUGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUUGAUUCGAAUUAUC-CUGGCUA ..((((.(((((..((((.((((((((((((..(-...((-(...)))...--)..)).)))).)))))).))))..))))).)))).............-....... ( -29.80) >consensus AAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGA_AAUGA_AUGAAUUCUC__GCGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUCGAACUGGC_UGG_UAA ...(((.(((((..((((.((((((((((((((.....................)))).)))).)))))).))))..))))).)))...................... (-25.71 = -25.27 + -0.44)


| Location | 11,920,048 – 11,920,151 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
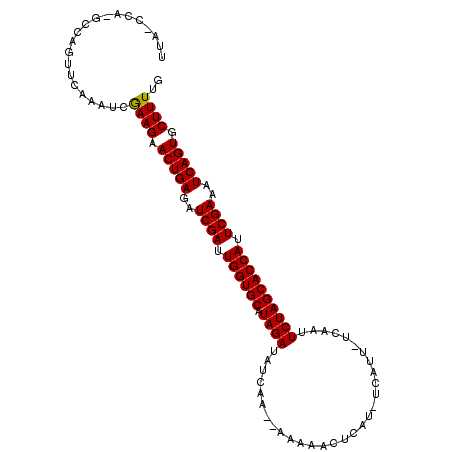
>3L_DroMel_CAF1 11920048 103 - 23771897 UUGCCCU-GCCACUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCAG--GAGAACCCAC-UCAAU-UUAACUCUAGCACCAUUCGAAAUCAGUGCUUUUG .......-((..(((((...))))).(((((..((((.((((((.((((......--(((......)-))...-.....)))))))))).))))..)))))))..... ( -32.24) >DroVir_CAF1 23888 104 - 1 GCA-CCA-GCCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCGCCCAAAAAGUAAU-UCAUU-UCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUU (((-(..-..((((((........))))))...((((.((((((.((((..................-.....-.....)))))))))).)))).....))))..... ( -29.59) >DroPse_CAF1 2703 104 - 1 UUACAAU-UACAGUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCAA--GAGGACUCGC-UAAUUUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUG .......-.............((((.(((((..((((.((((((.((((......--(((((.....-...)))))...)))))))))).))))..))))).)))).. ( -28.30) >DroGri_CAF1 12732 104 - 1 GUA-GCA-GCCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCGUCCAAGAAU-AAUUUAAUU-UCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUGU ..(-(((-..((((((........))))))...((((.((((((.((((..............-.........-.....)))))))))).))))......)))).... ( -27.59) >DroMoj_CAF1 14665 106 - 1 ACA-CCACACCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAACGCCCAAAAAGUCAUUUUAUU-UCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUU ...-.................((((.(((((..((((.((((((.((((.........((((....))))...-.....)))))))))).))))..))))).)))).. ( -27.43) >DroAna_CAF1 2225 103 - 1 UAGCCAG-GAUAAUUCGAAUCAAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCAA--GGAGAUCCGA-UCAUU-UCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUG .......-.............((((.(((((..((((.((((((.((((......--((....))((-.....-))...)))))))))).))))..))))).)))).. ( -30.50) >consensus UUA_CCA_GCCAGUUCAAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCAA__AAAAACUCAU_UCAUU_UCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUG .....................((((.(((((..((((.((((((.((((..............................)))))))))).))))..))))).)))).. (-26.86 = -26.73 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:16 2006