| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
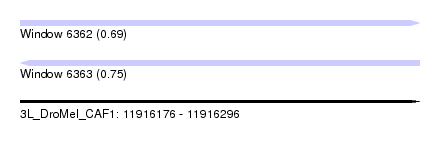
| Location | 11,916,176 – 11,916,296 |
| Length | 120 |
| Max. P | 0.750367 |

| Location | 11,916,176 – 11,916,296 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11916176 120 + 23771897 AAGGUCGUGAAUGCCGAGUACGAAAAGUCCGAGGACGAUCUGAAGGCUCUCCAGAGUGUGGGUCAGAUGGUCGGGGAAAUCCUGAAGCAGCUCACACCGGACAACUUCAUCGUGAAGGCC ............(((...(((((...(((....)))....(((((....(((.(.((((((((....((.(((((.....)))))..))))))))))))))...))))))))))..))). ( -39.10) >DroSec_CAF1 91978 120 + 1 AAGGUCGUGAAUGCCCAGUACGAGAAGUCCGAGGACGAUCUGAAAGCUCUCCAGAGUGUGGGUCAGAUGGUCGGGGAAGUUCUGAAGCAGCUCACCCCGGACAAUUUCAUCGUGAAGGCC ............(((...(((((((((((((..(((.((((((..((((....)))).....)))))).)))(((..(((((....).))))..))))))))...))).)))))..))). ( -44.10) >DroSim_CAF1 92238 120 + 1 AAGGUCGUGAAUGCCCAGUAUGAGAAGUCCGAGGACGAUCUGAAGGCUCUCCAGAGUGUGGGUCAGAUGGUCGGGGAAGUUCUGAAGCAGCUCACCCCGGACAACUUCAUCGUGAAGGCA ...........((((...(((((((((((((..(((.(((((((.((((....)))).)...)))))).)))(((..(((((....).))))..))))))))...))).)))))..)))) ( -42.50) >DroEre_CAF1 94428 120 + 1 AAGGUCGUGAAUGCCCAGUACGAGAAGUCCGAGGAGGACCUGAAGGCCCUCCAGAGUGUGGGUCAGAUGCUCGGGGAAAUCCUGAAGCAGCUCACCCCGGACAACUUCAUCGUGAAGGCC ............(((...(((((((((((((.(((((.((....)).))))).......((((.((.((((((((.....)))).)))).)).)))))))))...))).)))))..))). ( -48.10) >DroYak_CAF1 96529 120 + 1 AAGGUCGUGAAUGCCCAGUACGAGAAGUCCGAGGAGGAUCUGAAGGCGCUCCAGAGUGUGGGUCAGAUGCUCGGUGAAAUUCUGAAGCAGCUGACCCCGGACAACUUCAUCGUGAAGGCC ............(((...(((((((((((((.((((..((....))..)))).......(((((((.(((((((.......))).)))).))))))))))))...))).)))))..))). ( -44.80) >consensus AAGGUCGUGAAUGCCCAGUACGAGAAGUCCGAGGACGAUCUGAAGGCUCUCCAGAGUGUGGGUCAGAUGGUCGGGGAAAUUCUGAAGCAGCUCACCCCGGACAACUUCAUCGUGAAGGCC ............(((...((((((((((((...(((.((((((..((.((....)).))...)))))).)))((((.....(((...)))....))))))))...))).)))))..))). (-35.12 = -35.28 + 0.16)


| Location | 11,916,176 – 11,916,296 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11916176 120 - 23771897 GGCCUUCACGAUGAAGUUGUCCGGUGUGAGCUGCUUCAGGAUUUCCCCGACCAUCUGACCCACACUCUGGAGAGCCUUCAGAUCGUCCUCGGACUUUUCGUACUCGGCAUUCACGACCUU .(((...(((((((((((.(((((.(((...((..(((((...((...))...)))))..))))).))))).)).)))))....(((....)))...))))....)))............ ( -30.90) >DroSec_CAF1 91978 120 - 1 GGCCUUCACGAUGAAAUUGUCCGGGGUGAGCUGCUUCAGAACUUCCCCGACCAUCUGACCCACACUCUGGAGAGCUUUCAGAUCGUCCUCGGACUUCUCGUACUGGGCAUUCACGACCUU .((((..((((.(((...((((((((....(((...))).....))))(((.((((((......(((....)))...)))))).)))...)))))))))))...))))............ ( -37.70) >DroSim_CAF1 92238 120 - 1 UGCCUUCACGAUGAAGUUGUCCGGGGUGAGCUGCUUCAGAACUUCCCCGACCAUCUGACCCACACUCUGGAGAGCCUUCAGAUCGUCCUCGGACUUCUCAUACUGGGCAUUCACGACCUU (((((.....((((.(..((((((((....(((...))).....))))(((.((((((......(((....)))...)))))).)))...))))..)))))...)))))........... ( -36.70) >DroEre_CAF1 94428 120 - 1 GGCCUUCACGAUGAAGUUGUCCGGGGUGAGCUGCUUCAGGAUUUCCCCGAGCAUCUGACCCACACUCUGGAGGGCCUUCAGGUCCUCCUCGGACUUCUCGUACUGGGCAUUCACGACCUU .((((..((((.(((...(((((((((.((.(((((..((....))..))))).)).)))).......((((((((....)))))))).))))))))))))...))))............ ( -53.90) >DroYak_CAF1 96529 120 - 1 GGCCUUCACGAUGAAGUUGUCCGGGGUCAGCUGCUUCAGAAUUUCACCGAGCAUCUGACCCACACUCUGGAGCGCCUUCAGAUCCUCCUCGGACUUCUCGUACUGGGCAUUCACGACCUU .((((..((((.(((...((((((((((((.(((((..(.....)...))))).)))))))....(((((((...))))))).......))))))))))))...))))............ ( -43.90) >consensus GGCCUUCACGAUGAAGUUGUCCGGGGUGAGCUGCUUCAGAAUUUCCCCGACCAUCUGACCCACACUCUGGAGAGCCUUCAGAUCGUCCUCGGACUUCUCGUACUGGGCAUUCACGACCUU .((((..((((.(((...((((((((....(((...))).....))))(((.((((((((((.....))).).....)))))).)))...)))))))))))...))))............ (-31.56 = -31.56 + 0.00)

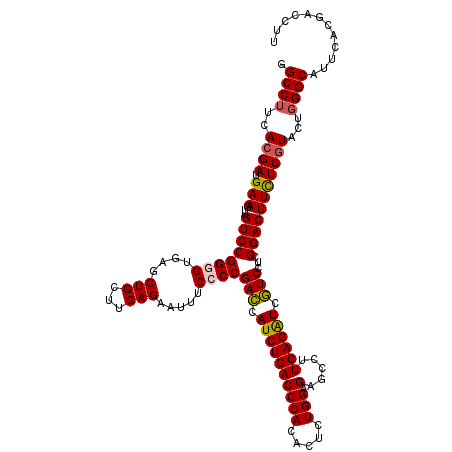

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:13 2006