| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,902,526 – 11,902,618 |
| Length | 92 |
| Max. P | 0.964370 |

| Location | 11,902,526 – 11,902,618 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11902526 92 + 23771897 A---AACAGACAUAUUUUGACACUUGGCCAAAAAAAGAAGAAAAAAGUCAAGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC .---..((((.....)))).(((((((((.................(((((.((...)).))))-)))))--.(((((........)))))))))).. ( -21.92) >DroSim_CAF1 78675 90 + 1 A---AACAGACACAUUUUGACACUUGGCC-AAAAAAGAUGAAA-AAAACAAGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC .---......(((...(..((((((.(.(-.............-.......).).))))))..)-.....--.(((((........)))))..))).. ( -20.55) >DroEre_CAF1 79952 75 + 1 A---AACAGACACAUUUUGACACUUGGCC-AAAA----------------AGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC .---......(((...(..((((((.(.(-....----------------.).).))))))..)-.....--.(((((........)))))..))).. ( -22.20) >DroYak_CAF1 81819 78 + 1 A---AACAGACACAUUUUGACACUUGGCC-AAAA----------------AGACGAAGUGUUGGGCCGCCAACACCCGAAAUCCAACGGGCAAGUGAC .---..((((.....)))).((((((.((-....----------------.......(((((((....)))))))..(.....)...)).)))))).. ( -21.50) >DroAna_CAF1 73001 93 + 1 AGGCACCAGACACUUUUUGACACUUGGCC-AAAAAUACAGAAA-UACAGAAAACUGUGUGUUGA-CGAGC--CGCCCGAAAUCCAGCGGGCAAGUGAC .(((.(..(((((((((((.(....)..)-)))))........-.((((....)))))))))..-.).))--)(((((........)))))....... ( -24.60) >consensus A___AACAGACACAUUUUGACACUUGGCC_AAAAA____GAAA__A___AAGACGAAGUGUUGG_CGGCU__CGCCCGAAAUCCAACGGGCAAGUGAC ..........(((...(..((((((.(..........................).))))))..).........(((((........)))))..))).. (-17.31 = -17.23 + -0.08)
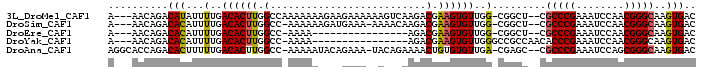


| Location | 11,902,526 – 11,902,618 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.51 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11902526 92 - 23771897 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCUUGACUUUUUUCUUCUUUUUUUGGCCAAGUGUCAAAAUAUGUCUGUU---U ((..((((((((........))))))--))..)-)..((((((.(((......................))).))))))...............---. ( -21.45) >DroSim_CAF1 78675 90 - 1 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCUUGUUUU-UUUCAUCUUUUUU-GGCCAAGUGUCAAAAUGUGUCUGUU---U ((..((((((((........))))))--))..)-)..((((((.(((.......-.............-))).))))))...............---. ( -21.55) >DroEre_CAF1 79952 75 - 1 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCU----------------UUUU-GGCCAAGUGUCAAAAUGUGUCUGUU---U ((..((((((((........))))))--))..)-)..((((((.(((.----------------....-))).))))))...............---. ( -23.20) >DroYak_CAF1 81819 78 - 1 GUCACUUGCCCGUUGGAUUUCGGGUGUUGGCGGCCCAACACUUCGUCU----------------UUUU-GGCCAAGUGUCAAAAUGUGUCUGUU---U (.((((((.(((..((((...(((((((((....))))))))).))))----------------...)-)).)))))).)..............---. ( -26.10) >DroAna_CAF1 73001 93 - 1 GUCACUUGCCCGCUGGAUUUCGGGCG--GCUCG-UCAACACACAGUUUUCUGUA-UUUCUGUAUUUUU-GGCCAAGUGUCAAAAAGUGUCUGGUGCCU .......(((((........)))))(--((...-(((((((((((....)))).-........(((((-(((.....)))))))))))).))).))). ( -27.30) >consensus GUCACUUGCCCGUUGGAUUUCGGGCG__AGCCG_CCAACACUUCGUCUU___U__UUUC____UUUUU_GGCCAAGUGUCAAAAUGUGUCUGUU___U (.(((..(((((........)))))............((((((.(((......................))).))))))......))).)........ (-16.23 = -16.51 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:09 2006