
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,193,290 – 1,193,381 |
| Length | 91 |
| Max. P | 0.738468 |

| Location | 1,193,290 – 1,193,381 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -19.10 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
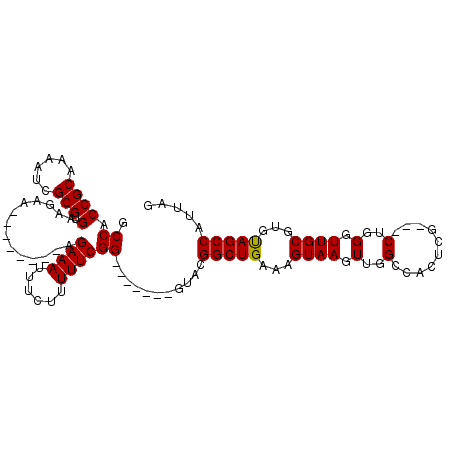
>3L_DroMel_CAF1 1193290 91 + 23771897 CUAAUGGCUGCACGCAACCCAG---CGAGUGGCCAACUUACUUUCAGCCAUAC-------CCGAAAAAGAAAUUUCU--------UUCUUCACGCGAUUUUGCGGUGGC ....((((..(.(((......)---)).)..))))...........(((((..-------..((((((((....)))--------)).))).((((....))))))))) ( -28.80) >DroSec_CAF1 8931 91 + 1 CUAAUGGCUACACGCAACCCAG---CGAGUGGCCAACUUACUUUCAGCCGUAC-------CCGAAAAAGAAAUUUCU--------UUCUUCACGCGAUUUUGCGGUGGC ....(((((((.(((......)---)).)))))))...........((((...-------..((((((((....)))--------)).))).((((....)))).)))) ( -27.30) >DroSim_CAF1 8326 94 + 1 CUAAUGGCUACACGCAACCCAGCGGCGAGUGGCCAACUUACUUUCAGCCGUAC-------CCGAAAAAGAAAUUUCU--------UUCUUCACGCGAUUUUGCGGUGGC ....(((((((.(((.........))).)))))))...........((((...-------..((((((((....)))--------)).))).((((....)))).)))) ( -26.20) >DroYak_CAF1 8450 106 + 1 CUAAUGGCUACACGCAACCCAG---UGAGUGGCCAACUUACUUUCAGCCUUACUCUUCGCCCGAAAAAGAAAUUUCUUUCUUUCUUUCUUCACGCGAUUUUGCGGUGGC ......(((((.(((((...((---(((((.....)))))))..............((((..((((((((((........))))))).)))..))))..)))))))))) ( -29.10) >consensus CUAAUGGCUACACGCAACCCAG___CGAGUGGCCAACUUACUUUCAGCCGUAC_______CCGAAAAAGAAAUUUCU________UUCUUCACGCGAUUUUGCGGUGGC ....(((((((.((...........)).)))))))...........(((.............((((......))))................((((....))))..))) (-19.10 = -18.73 + -0.38)



| Location | 1,193,290 – 1,193,381 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1193290 91 - 23771897 GCCACCGCAAAAUCGCGUGAAGAA--------AGAAAUUUCUUUUUCGG-------GUAUGGCUGAAAGUAAGUUGGCCACUCG---CUGGGUUGCGUGCAGCCAUUAG (((..(((......))).((((((--------(....)))))))....)-------))(((((((...((((.(..((.....)---)..).))))...)))))))... ( -32.80) >DroSec_CAF1 8931 91 - 1 GCCACCGCAAAAUCGCGUGAAGAA--------AGAAAUUUCUUUUUCGG-------GUACGGCUGAAAGUAAGUUGGCCACUCG---CUGGGUUGCGUGUAGCCAUUAG (((..(((......))).((((((--------(....)))))))....)-------))..(((((...((((.(..((.....)---)..).))))...)))))..... ( -30.00) >DroSim_CAF1 8326 94 - 1 GCCACCGCAAAAUCGCGUGAAGAA--------AGAAAUUUCUUUUUCGG-------GUACGGCUGAAAGUAAGUUGGCCACUCGCCGCUGGGUUGCGUGUAGCCAUUAG ((.(((((((..(.(((.((((((--------(....)))))))..(((-------((..(((..(.......)..)))))))).))).)..))))).)).))...... ( -30.10) >DroYak_CAF1 8450 106 - 1 GCCACCGCAAAAUCGCGUGAAGAAAGAAAGAAAGAAAUUUCUUUUUCGGGCGAAGAGUAAGGCUGAAAGUAAGUUGGCCACUCA---CUGGGUUGCGUGUAGCCAUUAG ((.(((((((..((((.(((.....((((((((....))))))))))).)))).((((..(((..(.......)..))))))).---.....))))).)).))...... ( -34.90) >consensus GCCACCGCAAAAUCGCGUGAAGAA________AGAAAUUUCUUUUUCGG_______GUACGGCUGAAAGUAAGUUGGCCACUCG___CUGGGUUGCGUGUAGCCAUUAG .((.((((......))).)..............((((......))))))...........(((((...((((.(..(..........)..).))))...)))))..... (-20.79 = -20.60 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:07 2006