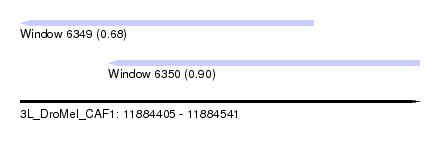
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,884,405 – 11,884,541 |
| Length | 136 |
| Max. P | 0.896160 |

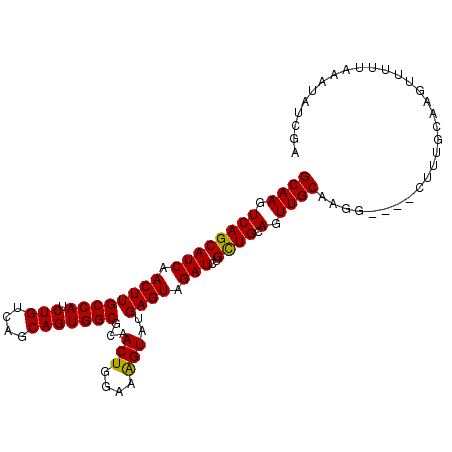
| Location | 11,884,405 – 11,884,505 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11884405 100 - 23771897 GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGGAAAGUAUGAGUAGAUCUGCUGCAGUUGCAAGG----CUUUGCAAGUUUUUAAAUGUCGA (((((((...........((((.(((....)))))))(((((((...((((.((.....)))))).)))))))..))----).))))................. ( -31.70) >DroSec_CAF1 59611 100 - 1 GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGGAAAGUAUGAGUAGAUCUGUUGCAGUUGCAAGG----CUUUGCAAGUUUUUAAAUUUCGA (((((((...........((((.(((....)))))))(((((((.((.....((.....))..)).)))))))..))----).))))................. ( -29.40) >DroSim_CAF1 60131 100 - 1 GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGAAAAGUAUGAGUAGAUCUGUUGCAGUUGCAAGG----CUUUGCAAGUUUUUAAAUUUCGA (((((((...........((((.(((....)))))))(((((((...((.((......))))....)))))))..))----).))))................. ( -28.10) >DroEre_CAF1 61714 98 - 1 GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGGAAAGUCUGAGUAGAUCCGCUGCAGUUGCAAGGUG------GCUGCUCCUGUAGUAUCGA ((((.((((((((.((((((((.(((....)))))))...(((....)))..)))).)))..)))).).))))..((((------.((((....)))))))).. ( -32.90) >DroYak_CAF1 62186 104 - 1 GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGGAAGGUCUGAGUAGAUCUGCUGCAGUUGCAAGGGAUACUCUACUUCACCUGAAAUAUCGA ((......))........((((.(((....)))))))(((((((..((((((....))))))....))))))).(((((.........)).))).......... ( -29.70) >consensus GCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCAACUGGAAAGUAUGAGUAGAUCUGCUGCAGUUGCAAGG____CUUUGCAAGUUUUUAAAUAUCGA ((((.((((((((.((((((((.(((....)))))))...(((....)))..)))).)))..)))).).))))............................... (-25.78 = -25.38 + -0.40)


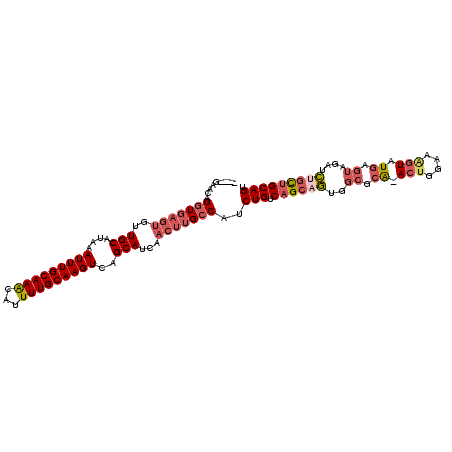
| Location | 11,884,435 – 11,884,541 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -28.00 |
| Energy contribution | -29.45 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11884435 106 - 23771897 ----GAACAGGUGAGUGUUGCAUAAAUUUGCAAACAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA-ACUGGAAAGUAUGAGUAGAUCUGCUGCAGU ----.....((..(((..(((....((((((((.....))))))))..)))...)))..))..(((.((((((..((.((-(((....))).)).))....))))))))). ( -37.20) >DroSec_CAF1 59641 106 - 1 ----GAACAGGUGAGUGUUGCAUAAAUUUGCAAACAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA-ACUGGAAAGUAUGAGUAGAUCUGUUGCAGU ----.....((..(((..(((....((((((((.....))))))))..)))...)))..))..(((.((((((..((.((-(((....))).)).))....))))))))). ( -34.80) >DroSim_CAF1 60161 106 - 1 ----GAACAGGUGAGUGUUGCAUAAAUUUGCAAACAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA-ACUGAAAAGUAUGAGUAGAUCUGUUGCAGU ----.....((..(((..(((....((((((((.....))))))))..)))...)))..))..(((.((((((..((.((-(((....))).)).))....))))))))). ( -34.80) >DroEre_CAF1 61742 106 - 1 ----GAACAGGUGAGUGUUGCAUAAAUUUGCAAGCAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA-ACUGGAAAGUCUGAGUAGAUCCGCUGCAGU ----((.(((((((((((((((......))).))))))..((((((........)))))))))))))).(((((((..(.-(((((.....)).))).)..)))))))... ( -36.10) >DroWil_CAF1 118551 103 - 1 UUCAGCUCAGAUGAGUGUUGCAUAAAUUUGCAAAUAUUUUGCAAGUCAGCAACA----GCCGCCUGCCAGCAAAUGCGCGCACCAUGAAUUCUUAC----UUCACGGCAGU ....((((....))))(((((....((((((((.....))))))))..))))).----((((..(((..((....))..)))...((((.......----))))))))... ( -27.50) >DroYak_CAF1 62220 106 - 1 ----GAGCAGGUGAGUGUUGCAUAAAUUUGCAAACAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA-ACUGGAAGGUCUGAGUAGAUCUGCUGCAGU ----(.((((((..((((((.....((((((((.....))))))))))))))..)))))))..(((.((((((..((.((-(((....))).)).))....))))))))). ( -36.30) >consensus ____GAACAGGUGAGUGUUGCAUAAAUUUGCAAACAUUUUGCAAGUCAGCAUCAACUUGCCAUCUGUCAGCAGUGGCGCA_ACUGGAAAGUAUGAGUAGAUCUGCUGCAGU .........(((((((..(((....(((((((((...)))))))))..)))...)))))))..(((.((((((..((.((.(((....))).)).))....))))))))). (-28.00 = -29.45 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:00 2006