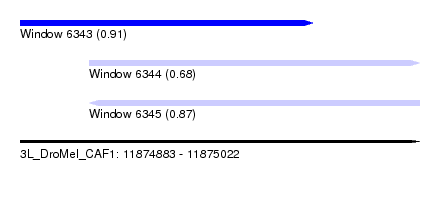
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,874,883 – 11,875,022 |
| Length | 139 |
| Max. P | 0.905209 |

| Location | 11,874,883 – 11,874,985 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.17 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
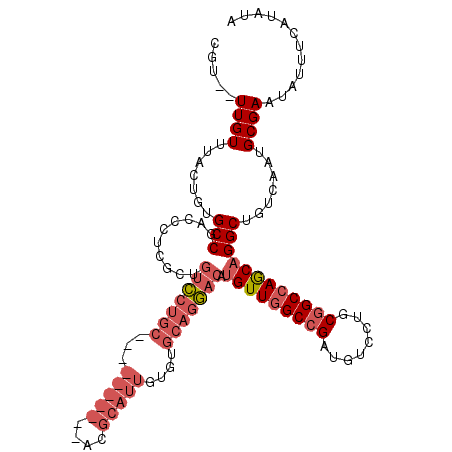
>3L_DroMel_CAF1 11874883 102 + 23771897 GACCACAGGGUCAAAGUUGUUUUCUAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCACACAAUGCGUCCGCACU ((((....))))..(((.((((((.....))))....(((((((...(((....))).(((((((((.(.....))))))))))...)))))))...))))) ( -36.10) >DroSec_CAF1 50331 86 + 1 GACCACAGGGUCAAAGUUGUUUUCUAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCAC---------------- (.((((((((((((.((...((((.....)))).....)).)))))..)))).))).)(((((((((.(.....))))))))))..---------------- ( -31.20) >DroSim_CAF1 50819 86 + 1 GACCACAGGGUCAAAGUUGUUUUCUAUAUGAAGUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCAC---------------- (.((((((((((((.((...((((.....)))).....)).)))))..)))).))).)(((((((((.(.....))))))))))..---------------- ( -30.90) >DroEre_CAF1 52387 101 + 1 GACCACGGGGUCAAAGUUGUUUUCUAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUC-GCCAACAUGUCCUGCACACAAUGCGUACGCACU ((((....))))..((((((((((.....))))....(((((((...(((....))).(((((((((.-......)))))))))...))))))).))).))) ( -35.40) >DroYak_CAF1 52254 102 + 1 GACCACAGGGUCAAAGUUGUUUUCUAUAUGGAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAACGGCCAACAUGUCCUGCACACAAUGCGUCCGCACU ((((....)))).................(((.....(((((((...(((....))).((((((((..........))))))))...))))))))))..... ( -34.80) >DroAna_CAF1 48812 94 + 1 GUCCGCAGGGUCAAAGUUGUUUUCUAUAUGAAAUAUUCGCAUUGACAGCCAGUUGGCCGCAGGACA-CGGCAAACAUGAACCGCACACUCGGCGC------- ((((((...(((((.((...((((.....)))).....)).))))).(((....))).)).)))).-..............(((.......))).------- ( -22.20) >consensus GACCACAGGGUCAAAGUUGUUUUCUAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCACACAAUGCGU_______ (.((((((((((((.((...((((.....)))).....)).)))))..)))).))).)(((((((((........))))))))).................. (-26.69 = -27.17 + 0.47)


| Location | 11,874,907 – 11,875,022 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.22 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11874907 115 + 23771897 UAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCACACAAUGCGUCCGCACUUCCGCAGGACAGCGAGGGUCGGCACAGUAAACAA--ACG ..............((((((...(((....))).(((((((((.(.....))))))))))...))))))(((((.(((..(((......)))..)))))).)).........--... ( -42.80) >DroSec_CAF1 50355 96 + 1 UAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCAC-------------------GCAGGACAGCGAGGGUCGGCACAGUAAACAA--ACG .................(((...((.(((((((((((((((((.(.....)))))))))).(-------------------((......)))..))))))))..))...)))--... ( -33.10) >DroSim_CAF1 50843 96 + 1 UAUAUGAAGUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCAC-------------------GCAGGACAGCGAGGGUCGGCACAGUAAACAA--ACG .................(((...((.(((((((((((((((((.(.....)))))))))).(-------------------((......)))..))))))))..))...)))--... ( -33.10) >DroEre_CAF1 52411 114 + 1 UAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUC-GCCAACAUGUCCUGCACACAAUGCGUACGCACUUCCGCAGGACGACGAGGGUAGGCACAGUAAACAA--ACG .............(((((((...(((....))).(((((((((.-......)))))))))...)))))))...((.(((((...(.....)..))).))))...........--... ( -34.70) >DroYak_CAF1 52278 115 + 1 UAUAUGGAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAACGGCCAACAUGUCCUGCACACAAUGCGUCCGCACUUCCGCAGGACAACGAGGGUAGGCACAGUAAACAA--ACG ....(((..(((((((((((...(((....))).((((((((..........))))))))...))))))((((((......)).)))).....)))))..).))........--... ( -38.80) >DroAna_CAF1 48836 102 + 1 UAUAUGAAAUAUUCGCAUUGACAGCCAGUUGGCCGCAGGACA-CGGCAAACAUGAACCGCACACUCGGCGC--------------CAGAACGAGGGUAGGCCCGUCAAACAACAACA .................(((((.(((.(((.((((.......-)))).))).......((...((((....--------------.....)))).)).)))..)))))......... ( -24.10) >consensus UAUAUGAAAUAUUCGCAUUGACAGCCUGCUGGCCGCAGGACAUCGGCCAACAUGUCCUGCACACAAUGCGU__________GCAGGACAACGAGGGUAGGCACAGUAAACAA__ACG .......................(((.((((((((........)))))....((((((((.....................)))))))).....))).)))................ (-21.99 = -23.22 + 1.22)

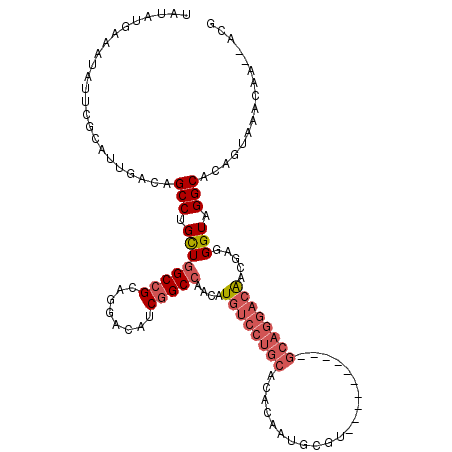
| Location | 11,874,907 – 11,875,022 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -28.54 |
| Energy contribution | -30.87 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.865055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11874907 115 - 23771897 CGU--UUGUUUACUGUGCCGACCCUCGCUGUCCUGCGGAAGUGCGGACGCAUUGUGUGCAGGACAUGUUGGCCGAUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUAUUUCAUAUA .((--((((.......(((.........(((((((((.((((((....))))).).)))))))))(((((((((........)))))))))))).......)))))).......... ( -46.44) >DroSec_CAF1 50355 96 - 1 CGU--UUGUUUACUGUGCCGACCCUCGCUGUCCUGC-------------------GUGCAGGACAUGUUGGCCGAUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUAUUUCAUAUA .((--((((.......(((.........(((((((.-------------------...)))))))(((((((((........)))))))))))).......)))))).......... ( -37.04) >DroSim_CAF1 50843 96 - 1 CGU--UUGUUUACUGUGCCGACCCUCGCUGUCCUGC-------------------GUGCAGGACAUGUUGGCCGAUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUACUUCAUAUA .((--((((.......(((.........(((((((.-------------------...)))))))(((((((((........)))))))))))).......)))))).......... ( -37.04) >DroEre_CAF1 52411 114 - 1 CGU--UUGUUUACUGUGCCUACCCUCGUCGUCCUGCGGAAGUGCGUACGCAUUGUGUGCAGGACAUGUUGGC-GAUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUAUUUCAUAUA .((--((((.......(((......(((.((((((((.((((((....))))).).)))))))))))..)))-((((.(((((.....))))).))))...)))))).......... ( -40.00) >DroYak_CAF1 52278 115 - 1 CGU--UUGUUUACUGUGCCUACCCUCGUUGUCCUGCGGAAGUGCGGACGCAUUGUGUGCAGGACAUGUUGGCCGUUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUAUUCCAUAUA .((--((((.......((((........(((((((((.((((((....))))).).))))))))).(((((((((......))))))))))))).......)))))).......... ( -48.44) >DroAna_CAF1 48836 102 - 1 UGUUGUUGUUUGACGGGCCUACCCUCGUUCUG--------------GCGCCGAGUGUGCGGUUCAUGUUUGCCG-UGUCCUGCGGCCAACUGGCUGUCAAUGCGAAUAUUUCAUAUA ..((((...((((((((((.((....))...)--------------))((((((..((((((........))))-))..)).)))).......))))))).))))............ ( -31.90) >consensus CGU__UUGUUUACUGUGCCGACCCUCGCUGUCCUGC__________ACGCAUUGUGUGCAGGACAUGUUGGCCGAUGUCCUGCGGCCAGCAGGCUGUCAAUGCGAAUAUUUCAUAUA .....((((.......(((..........(((((((...(((((....)))))....))))))).(((((((((........)))))))))))).......))))............ (-28.54 = -30.87 + 2.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:55 2006