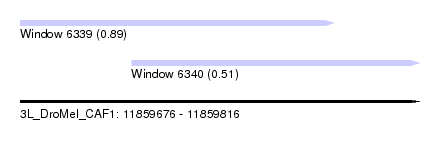
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,859,676 – 11,859,816 |
| Length | 140 |
| Max. P | 0.891336 |

| Location | 11,859,676 – 11,859,786 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11859676 110 + 23771897 UUCUCAGGCUCUUGCCGUAUUUUUUUUUUCCACCCAUUUCCCAUGCUCUGCGGCGGAAAAAGUUUUUUCAGCUUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAA .....((((((..((.(......((((((((.((((............)).)).))))))))......).))....)))))).(((((.......))))).......... ( -24.50) >DroSec_CAF1 35575 87 + 1 UUCUCUGCCUCUUGCUGUAAU-----------------------GCUCUGCGGCGGAAAAAGUUUUUCCAGCUUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAA ...((((((.(..((......-----------------------))...).))))))..(((((.....)))))..((((...(((((.......)))))....)))).. ( -22.50) >DroSim_CAF1 36038 108 + 1 UUCUCAGCCUCUUGCUGUAAUUU--UUUCCCUCCCAUUUCCCGCGCUCUGCGGCGGAAAAAGUUUUUCCAGCCUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAA ......((.....((((.(((((--(((((..........((((.....)))).))))))))))....))))..))((((...(((((.......)))))....)))).. ( -26.40) >DroEre_CAF1 38049 94 + 1 UGCUCAGCCUCUUGCACUCAUUU--UG-UCCACAAUUUCGCCAUGCUCUGCGGCGGAAAAAGUUUUUCCA-------------GGCAACUUUCUUUUGCCAUUCACUCAA ......(((.(..(((......(--((-....)))........)))...).)))(((((.....))))).-------------(((((.......))))).......... ( -18.04) >DroYak_CAF1 36841 94 + 1 UUCUCAGCCUCUUGCACUCAUUU--UU-UUCACAAUUUUCCCAUGCUUUGCGGCGGAAAAAGUUUUUCCC-------------AGCAACUUUCUUUUGCCAUUCAUUCGA ......((.....(((.......--..-...............)))...))(((((((((((((......-------------...)))))).))))))).......... ( -11.46) >consensus UUCUCAGCCUCUUGCAGUAAUUU__UU_UCCACAAAUUUCCCAUGCUCUGCGGCGGAAAAAGUUUUUCCAGC_UGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAA ......(((.(..((.............................))...).)))(((((.....)))))..............(((((.......))))).......... (-11.33 = -11.57 + 0.24)

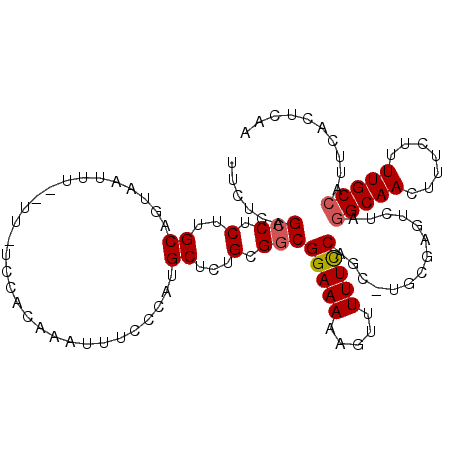
| Location | 11,859,715 – 11,859,816 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11859715 101 + 23771897 CCCAUGCUCUGCGGCGGAAAAAGUUUUUUCAGCUUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAAUGCUUUC--UCUCCAGUAGGGCA--CAAAGCCGA ....((((((((((.(((.(((((.......(....)((((...(((((.......)))))....))))...))))).--))))).))))))))--......... ( -27.00) >DroSec_CAF1 35596 98 + 1 -----GCUCUGCGGCGGAAAAAGUUUUUCCAGCUUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAAUGCUUUC--UCUGUAGUAGGGCACACAAAGCCGA -----(((((((.(((((.(((((.......(....)((((...(((((.......)))))....))))...))))).--))))).)))))))............ ( -29.50) >DroSim_CAF1 36075 103 + 1 CCCGCGCUCUGCGGCGGAAAAAGUUUUUCCAGCCUGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAAUGCUUUC--UCUCCAGUGGGGCACACAAAGCCGA .((((((((.((((((((((.....))))).))).))))))...(((((.......))))).................--......))))(((.......))).. ( -31.00) >DroEre_CAF1 38085 88 + 1 GCCAUGCUCUGCGGCGGAAAAAGUUUUUCCA-------------GGCAACUUUCUUUUGCCAUUCACUCAAUGCCUUC--UCCUCAGUGGGGCA--CAAAGCCGA ...........(((((((((.....))))).-------------(((((.......)))))..........(((((((--(....)).))))))--....)))). ( -23.40) >DroYak_CAF1 36877 90 + 1 CCCAUGCUUUGCGGCGGAAAAAGUUUUUCCC-------------AGCAACUUUCUUUUGCCAUUCAUUCGAUGCUUUCACUCCACAGUUGGGCA--CAAAGCCGA .....((((((.(((((((((((((......-------------...)))))).)))))))..........(((((..(((....))).)))))--))))))... ( -21.10) >consensus CCCAUGCUCUGCGGCGGAAAAAGUUUUUCCAGC_UGCGAGUCUAGGCAACUUUCUUUUGCCAUUCACUCAAUGCUUUC__UCUCCAGUAGGGCA__CAAAGCCGA .....(((((((((((((((.....)))))..............(((((.......)))))...........)))...........)))))))............ (-18.86 = -18.62 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:50 2006