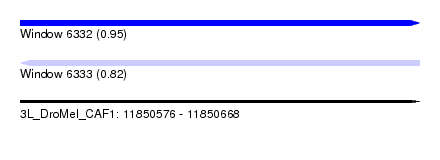
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,850,576 – 11,850,668 |
| Length | 92 |
| Max. P | 0.946849 |

| Location | 11,850,576 – 11,850,668 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11850576 92 + 23771897 UGCACCA-AAGGCACCUGGACCUGGCCUGCAUU-CC--UUGUAUAUCGAGCAAAUGUCAGGCCGAAAGGUCAACGAACCCAGGUGAACACAGAAAG- .......-....(((((((((((((((((((((-.(--(((.....))))..)))).))))))...))))........)))))))...........- ( -30.70) >DroSec_CAF1 26596 90 + 1 UGCACCA-AAGGCACCUGGACCUGGCCUGCAUU-CC--UUGUAUACCAAGCAAAUGUCAGGCCGAAAGGUCAACGAACCUAGGUGAAC--AGAAAG- ..((((.-.(((......(((((((((((((((-.(--(((.....))))..)))).))))))...)))))......))).))))...--......- ( -31.20) >DroEre_CAF1 25984 89 + 1 UGCACCA-GAGGCACCUGGUCCUGGCCCGCAUU-CC--UUGCAUGGCGAGCAAAUGUCAGGCCGAAAGGUCAACGCACCCAGGUGC-C--CGAAAG- .......-(.(((((((((..((.(((.(((..-..--.)))..))).)).........((((....)))).......))))))))-)--).....- ( -37.60) >DroYak_CAF1 27414 89 + 1 UGCACCA-GAGGCACCUGGACCUGGCCUGCAUU-CC--UUGUACGGCGAGCAAAUGUCAGGCCGAAAGGUCAACGAACCCAGGUGC-C--UGAAAG- .......-.((((((((((..((.(((((((..-..--.)))).))).)).........((((....)))).......))))))))-)--).....- ( -40.00) >DroAna_CAF1 26569 92 + 1 UGUAAAA-AAGGCACAUGGACCUGACUUGCAUU-GCUGUUGCAUGAAGAGCAAAUGUCAGGGCGAAAGGUCACCGAACCCAGGUGC-C--ACAAGCC .......-..(((((.(((.((((((.....((-(((.((.....)).)))))..)))))).(....)..........))).))))-)--....... ( -30.20) >DroPer_CAF1 25785 89 + 1 GGGGCCACGUGGCACAUGGACCUGGUCUGCACUGCC--GUGCAUGACAAGCAGGUGCCAAGAGGAAAGGUCACCGAACCCAGCUGA-----GGAGG- ((((((....)))...((((((((((((((((....--))))).)))...))))).)))...((........))...)))......-----.....- ( -30.00) >consensus UGCACCA_AAGGCACCUGGACCUGGCCUGCAUU_CC__UUGCAUGACGAGCAAAUGUCAGGCCGAAAGGUCAACGAACCCAGGUGA_C__AGAAAG_ ............(((((((((((((((((((((......(((.......))))))).))))))...))))........)))))))............ (-20.05 = -20.50 + 0.45)



| Location | 11,850,576 – 11,850,668 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -18.66 |
| Energy contribution | -20.69 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11850576 92 - 23771897 -CUUUCUGUGUUCACCUGGGUUCGUUGACCUUUCGGCCUGACAUUUGCUCGAUAUACAA--GG-AAUGCAGGCCAGGUCCAGGUGCCUU-UGGUGCA -...........((((.((((.(.(.(((((...((((((.(((((.((.(.....).)--))-))))))))))))))).).).)))).-.)))).. ( -32.50) >DroSec_CAF1 26596 90 - 1 -CUUUCU--GUUCACCUAGGUUCGUUGACCUUUCGGCCUGACAUUUGCUUGGUAUACAA--GG-AAUGCAGGCCAGGUCCAGGUGCCUU-UGGUGCA -......--...((((.((((.(.(.(((((...((((((.(((((.((((.....)))--))-))))))))))))))).).).)))).-.)))).. ( -37.90) >DroEre_CAF1 25984 89 - 1 -CUUUCG--G-GCACCUGGGUGCGUUGACCUUUCGGCCUGACAUUUGCUCGCCAUGCAA--GG-AAUGCGGGCCAGGACCAGGUGCCUC-UGGUGCA -.....(--(-((((((((.......((....))((((((.(((((.((.((...)).)--))-))))))))))....)))))))))).-....... ( -36.60) >DroYak_CAF1 27414 89 - 1 -CUUUCA--G-GCACCUGGGUUCGUUGACCUUUCGGCCUGACAUUUGCUCGCCGUACAA--GG-AAUGCAGGCCAGGUCCAGGUGCCUC-UGGUGCA -......--.-(((((.((((.(.(.(((((...((((((.(((((.((.(.....).)--))-))))))))))))))).).).)))).-.))))). ( -37.00) >DroAna_CAF1 26569 92 - 1 GGCUUGU--G-GCACCUGGGUUCGGUGACCUUUCGCCCUGACAUUUGCUCUUCAUGCAACAGC-AAUGCAAGUCAGGUCCAUGUGCCUU-UUUUACA (((.(((--(-(.(((((.(((.(((((....)))))..)))((((((......(((....))-)..)))))))))))))))).)))..-....... ( -30.20) >DroPer_CAF1 25785 89 - 1 -CCUCC-----UCAGCUGGGUUCGGUGACCUUUCCUCUUGGCACCUGCUUGUCAUGCAC--GGCAGUGCAGACCAGGUCCAUGUGCCACGUGGCCCC -.....-----......(((...(....)....((.(.((((((..(((((((.(((((--....))))))).)))))....)))))).).))))). ( -29.70) >consensus _CUUUCU__G_GCACCUGGGUUCGUUGACCUUUCGGCCUGACAUUUGCUCGCCAUACAA__GG_AAUGCAGGCCAGGUCCAGGUGCCUC_UGGUGCA ............((((.(((..(.(.(((((...((((((.((((...................))))))))))))))).).)..)))...)))).. (-18.66 = -20.69 + 2.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:43 2006