
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,843,914 – 11,844,013 |
| Length | 99 |
| Max. P | 0.860403 |

| Location | 11,843,914 – 11,844,013 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.95 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11843914 99 + 23771897 CUACGUCGAGGACUUACGGCUCGAGUGUCU----CCCUGG------UCCUUGAUGCCAUUCACCC----AGAAUUCCGCCCAGAAUCUGCUCCUUCGCAGUCACCC-CGCAAAA ...(((((((((((...(((......))).----....))------)))))))))..((((....----.))))...((.......((((......))))......-.)).... ( -25.24) >DroPse_CAF1 18689 89 + 1 CCUCGUCGAGAGCUUACGGCUCGAGUGUCUGUUUCUCCGG------CCCUCGAUGCCAUUCACUC------------------UAUCUGCUCUUUCGCAGUCCCCC-AGCCCCA ........((((.....((((((((.(((.(.....).))------).))))).))).....)))------------------)..((((......))))......-....... ( -22.30) >DroEre_CAF1 19541 98 + 1 CCACGUCGAGGGCUUACGGCUCGAGUGUCU----CUCUGG------UCCUCGAUGCCAUUCACCC----AGAAUUCCGCCCAGAAUCUGCUCUUUCGCAGUCCCUC-C-CCACA .........((((....((((((((..((.----....))------..))))).)))((((....----.))))...)))).....((((......))))......-.-..... ( -28.00) >DroYak_CAF1 20618 105 + 1 CCACGUCGAGGGCUUACGGCUCGAGUGUCU----CUCUGGUCCUGGUCCUCGAUGCCAUUCACCC----AGAAUUCCGCCCACAAUCUGCUCUUUCGCAGUCUCCCGC-CCCCA ...(((((((((((...((((.(((.....----))).))))..)))))))))))..((((....----.))))...((.......((((......))))......))-..... ( -31.22) >DroAna_CAF1 18678 88 + 1 CCUCGUCGAGGGCUUACGACUCGAGUGUCU----UUCGGG------CCCUCGAUGCCAUUCAGCCCGGAAGAAUG------GGUAUCUGCUCUUUCGCAGUCCC---------- ...((((((((((((..(((......))).----...)))------))))))))).......(((((......))------)))..((((......))))....---------- ( -33.50) >DroPer_CAF1 18697 89 + 1 CCUCGUCGAGAGCUUACGGCUCGAGUGUCUGUUUCUCCGG------CCCUCGAUGCCAUUCACUC------------------UAUCUGCUCUUUCGCAGUCCCCC-AGCCCCA ........((((.....((((((((.(((.(.....).))------).))))).))).....)))------------------)..((((......))))......-....... ( -22.30) >consensus CCACGUCGAGGGCUUACGGCUCGAGUGUCU____CUCUGG______CCCUCGAUGCCAUUCACCC____AGAAUU______AGAAUCUGCUCUUUCGCAGUCCCCC_C_CCCCA ...(((((((((.......((.(((.........))).))......)))))))))...............................((((......)))).............. (-16.12 = -15.95 + -0.17)

| Location | 11,843,914 – 11,844,013 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -23.21 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
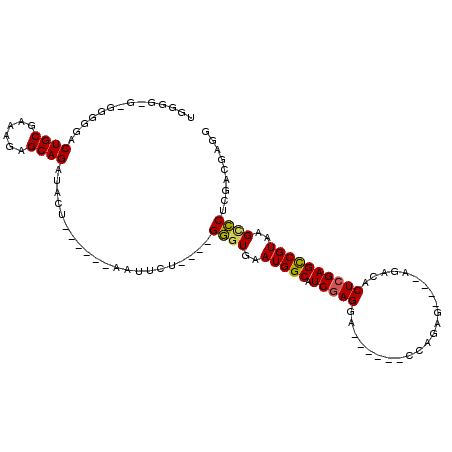
>3L_DroMel_CAF1 11843914 99 - 23771897 UUUUGCG-GGGUGACUGCGAAGGAGCAGAUUCUGGGCGGAAUUCU----GGGUGAAUGGCAUCAAGGA------CCAGGG----AGACACUCGAGCCGUAAGUCCUCGACGUAG ...((((-(((((.((((......)))).((((((.(((....))----)((((.....)))).....------))))))----...)))))(((.(....)..)))..)))). ( -30.60) >DroPse_CAF1 18689 89 - 1 UGGGGCU-GGGGGACUGCGAAAGAGCAGAUA------------------GAGUGAAUGGCAUCGAGGG------CCGGAGAAACAGACACUCGAGCCGUAAGCUCUCGACGAGG .....((-.(..((((((......))))...------------------((((..(((((.(((((..------(.(......).)...))))))))))..))))))..).)). ( -29.70) >DroEre_CAF1 19541 98 - 1 UGUGG-G-GAGGGACUGCGAAAGAGCAGAUUCUGGGCGGAAUUCU----GGGUGAAUGGCAUCGAGGA------CCAGAG----AGACACUCGAGCCGUAAGCCCUCGACGUGG ...(.-.-(((((.((((......))))......(((((..((((----.((((.....)))).))))------)).(((----.....)))..))).....)))))..).... ( -32.60) >DroYak_CAF1 20618 105 - 1 UGGGG-GCGGGAGACUGCGAAAGAGCAGAUUGUGGGCGGAAUUCU----GGGUGAAUGGCAUCGAGGACCAGGACCAGAG----AGACACUCGAGCCGUAAGCCCUCGACGUGG .((((-((......((((......))))......(((((..((((----((.(((......)))....)))))))).(((----.....)))..)))....))))))....... ( -35.40) >DroAna_CAF1 18678 88 - 1 ----------GGGACUGCGAAAGAGCAGAUACC------CAUUCUUCCGGGCUGAAUGGCAUCGAGGG------CCCGAA----AGACACUCGAGUCGUAAGCCCUCGACGAGG ----------(((.((((......))))...))------).(((.((.(((((..(((((.(((((..------..(...----.)...)))))))))).)))))..)).))). ( -35.00) >DroPer_CAF1 18697 89 - 1 UGGGGCU-GGGGGACUGCGAAAGAGCAGAUA------------------GAGUGAAUGGCAUCGAGGG------CCGGAGAAACAGACACUCGAGCCGUAAGCUCUCGACGAGG .....((-.(..((((((......))))...------------------((((..(((((.(((((..------(.(......).)...))))))))))..))))))..).)). ( -29.70) >consensus UGGGG_G_GGGGGACUGCGAAAGAGCAGAUACU______AAUUCU____GGGUGAAUGGCAUCGAGGA______CCAGAG____AGACACUCGAGCCGUAAGCCCUCGACGAGG ..............((((......)))).....................((((..(((((.(((((.......................))))))))))..))))......... (-23.21 = -22.85 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:41 2006