| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
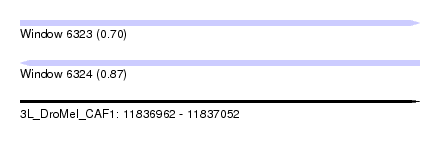
| Location | 11,836,962 – 11,837,052 |
| Length | 90 |
| Max. P | 0.869504 |

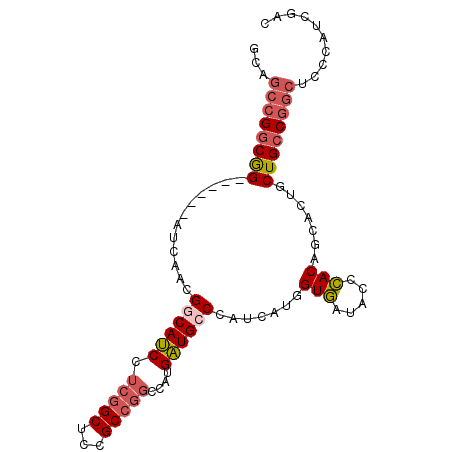
| Location | 11,836,962 – 11,837,052 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11836962 90 + 23771897 GUCGAUGGGAGCCGGCAGCAGUGCUGUGGGUAUCACCAUGAUAGGCAUCAUGGCCGGCGGAGCCGAGGAUGUCGUUGAU------CCGCCGGCUGC (((.(((((((((.(((((...))))).))).)).))))....))).....((((((((((..(((.(....).))).)------))))))))).. ( -44.40) >DroSec_CAF1 14306 90 + 1 GUCGAUGGAUGCCGGCAGCAGUGCUGUGGGUAUCACCAUGAUGGGCAUCAUGGCCGGCGGAGCCGAGGAUGCCGUUGAU------CCGCCGGCUGC ...((((((((((.(((((...))))).)))))).((.....)).))))..((((((((((..(((.(....).))).)------))))))))).. ( -47.80) >DroSim_CAF1 13845 90 + 1 GUCGAUGGGUGCCGGCAGCAGUGCUGUGGGUAUCACCAUGAUGGGCAUCAUGGCCGGCGGAGCCGAGGAUGCCGUUGAU------CCGCCGGCUGC ...((((((((((.(((((...))))).)))))).((.....)).))))..((((((((((..(((.(....).))).)------))))))))).. ( -47.20) >DroEre_CAF1 13685 90 + 1 GUCUAUGGCAGCCGGCAGAAGUGCUGUGGGUAUCACCAUGAUGGGCAUCAUGGCCGGCGGAGCCGAGGAUGACGCUGAU------CCGCCGGCUGC .......(((((((((....((.((((.((.....(((((((....))))))))).)))).))...((((.......))------))))))))))) ( -40.70) >DroYak_CAF1 14461 90 + 1 GUCGAUGGGAGCCGGCAGAAGAGCUGUGGGUAUCACCAUGAUGGGCAUCAUGGCCGGCGCAGCCGAGGAUGCCGUUGCU------CCGCCGGCUGC ((((...((.(((.((((.....)))).))).)).(((((((....))))))).))))(((((((.(((.((....)))------))..))))))) ( -43.20) >DroMoj_CAF1 31808 93 + 1 CGCUCCGGGC---GCCAGCAGCGCUGUCGGUAUGACCAUUAUGGGCAACAUUGUUGGCGCCGCUGCCGUUGCCGGCUGUGUGUUGCUGCCGCACGG ....(((.((---(.(((((((((.(((((((((.((.....)).))........((((....))))..)))))))...))))))))).))).))) ( -41.30) >consensus GUCGAUGGGAGCCGGCAGCAGUGCUGUGGGUAUCACCAUGAUGGGCAUCAUGGCCGGCGGAGCCGAGGAUGCCGUUGAU______CCGCCGGCUGC ((((.(((..(((.((((.....)))).)))............((((((.....((((...))))..))))))............))).))))... (-26.80 = -26.92 + 0.12)

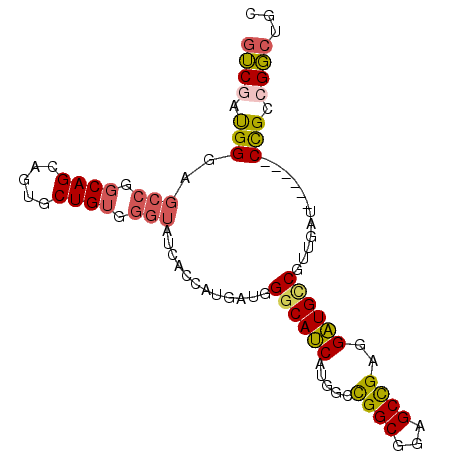

| Location | 11,836,962 – 11,837,052 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11836962 90 - 23771897 GCAGCCGGCGG------AUCAACGACAUCCUCGGCUCCGCCGGCCAUGAUGCCUAUCAUGGUGAUACCCACAGCACUGCUGCCGGCUCCCAUCGAC (((((((((((------(....(((.....)))..)))))))((((((((....))))))))...............))))).............. ( -33.40) >DroSec_CAF1 14306 90 - 1 GCAGCCGGCGG------AUCAACGGCAUCCUCGGCUCCGCCGGCCAUGAUGCCCAUCAUGGUGAUACCCACAGCACUGCUGCCGGCAUCCAUCGAC ...((((((((------.......((.....((((...))))((((((((....))))))))..........))....)))))))).......... ( -35.60) >DroSim_CAF1 13845 90 - 1 GCAGCCGGCGG------AUCAACGGCAUCCUCGGCUCCGCCGGCCAUGAUGCCCAUCAUGGUGAUACCCACAGCACUGCUGCCGGCACCCAUCGAC ...((((((((------.......((.....((((...))))((((((((....))))))))..........))....)))))))).......... ( -35.60) >DroEre_CAF1 13685 90 - 1 GCAGCCGGCGG------AUCAGCGUCAUCCUCGGCUCCGCCGGCCAUGAUGCCCAUCAUGGUGAUACCCACAGCACUUCUGCCGGCUGCCAUAGAC (((((((((((------(...((........((((...))))((((((((....))))))))..........))...))))))))))))....... ( -44.40) >DroYak_CAF1 14461 90 - 1 GCAGCCGGCGG------AGCAACGGCAUCCUCGGCUGCGCCGGCCAUGAUGCCCAUCAUGGUGAUACCCACAGCUCUUCUGCCGGCUCCCAUCGAC ((((((((.((------(((....)).)))))))))))((((((((((((....))))))(((.....))).........)))))).......... ( -42.80) >DroMoj_CAF1 31808 93 - 1 CCGUGCGGCAGCAACACACAGCCGGCAACGGCAGCGGCGCCAACAAUGUUGCCCAUAAUGGUCAUACCGACAGCGCUGCUGGC---GCCCGGAGCG (((.(((.(((((.(.....((((....)))).((((((.((....)).)))).......(((.....))).))).))))).)---)).))).... ( -40.20) >consensus GCAGCCGGCGG______AUCAACGGCAUCCUCGGCUCCGCCGGCCAUGAUGCCCAUCAUGGUGAUACCCACAGCACUGCUGCCGGCUCCCAUCGAC ...((((((((............((((((.(((((...)))))....)))))).......(((.....))).......)))))))).......... (-25.64 = -26.45 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:35 2006