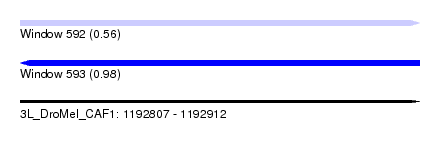
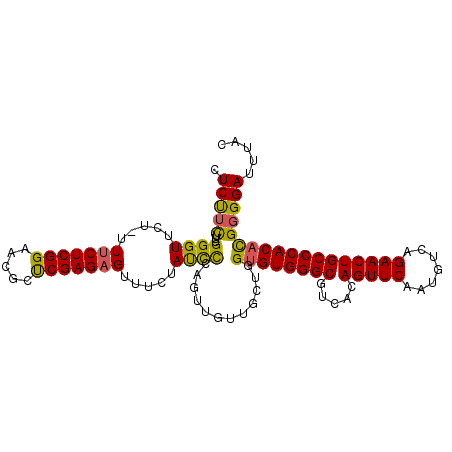
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,192,807 – 1,192,912 |
| Length | 105 |
| Max. P | 0.976982 |

| Location | 1,192,807 – 1,192,912 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -24.10 |
| Energy contribution | -25.16 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1192807 105 + 23771897 GUAAAUCCCCGUGUGGGCAGUUCUGACAUUGAACUGUGACGCCCACACCAUCAACAACUGGGAUAGAAACUCUCGAGCGUUCCGAGCAA-AGAACCCAACGAAGAG ....(((((.(((((((((((((.......))))).....))))))))...........))))).....((((((...((((.......-.))))....)).)))) ( -29.00) >DroSec_CAF1 8454 105 + 1 GUAAAUCCCCGUGUGGGCAGUUCUGACAUUGAACUGUGACGCCCACACCAACAACAACUGGGAUAGAAACUCUCGAGCGUUCCGAGAAA-AGAACCCAAAGAAGAG ..........(((((((((((((.......))))).....))))))))..........((((........(((((.......)))))..-....))))........ ( -28.34) >DroSim_CAF1 7844 105 + 1 GUAAAUCCCCGUGUGGGCAGUUCUGACAUUGAACUGUGACGCCCACACCAGCAACAACUGGGAUAGAAACUCUCGAGCGUUCCGAGAAA-AGAACCCAAAGAGGAG .....(((.((((((((((((((.......))))).....))))))))..........((((........(((((.......)))))..-....))))..).))). ( -31.64) >DroYak_CAF1 7912 102 + 1 GUAAAUCCCCAUGUGGGCAGUUCUGACAUUGAACUGUGACGCCCACGCCAGC---AACUUAAGUAGAAACUCUCGGGUAUUCCGAGAAAAAGAACCCAUAA-AGAA ............(((((((((((.......))))).....))))))......---...............((((((.....))))))..............-.... ( -23.80) >consensus GUAAAUCCCCGUGUGGGCAGUUCUGACAUUGAACUGUGACGCCCACACCAGCAACAACUGGGAUAGAAACUCUCGAGCGUUCCGAGAAA_AGAACCCAAAGAAGAG ..........(((((((((((((.......))))).....))))))))..........((((........(((((.......))))).......))))........ (-24.10 = -25.16 + 1.06)

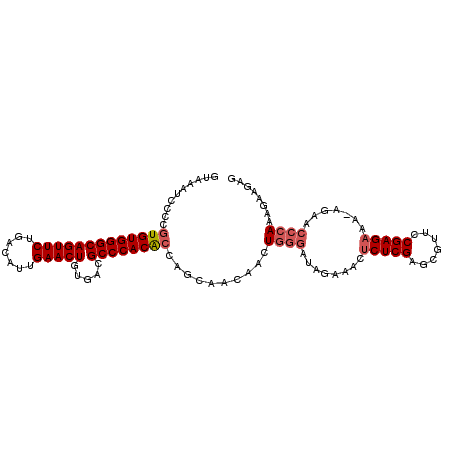
| Location | 1,192,807 – 1,192,912 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -32.04 |
| Energy contribution | -31.48 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1192807 105 - 23771897 CUCUUCGUUGGGUUCU-UUGCUCGGAACGCUCGAGAGUUUCUAUCCCAGUUGUUGAUGGUGUGGGCGUCACAGUUCAAUGUCAGAACUGCCCACACGGGGAUUUAC .(((((.(((((....-..((((.((....))..))))......))))).........((((((((.....(((((.......))))))))))))))))))..... ( -34.20) >DroSec_CAF1 8454 105 - 1 CUCUUCUUUGGGUUCU-UUUCUCGGAACGCUCGAGAGUUUCUAUCCCAGUUGUUGUUGGUGUGGGCGUCACAGUUCAAUGUCAGAACUGCCCACACGGGGAUUUAC .(((((.(((((....-.(((((((.....))))))).......))))).........((((((((.....(((((.......))))))))))))))))))..... ( -35.50) >DroSim_CAF1 7844 105 - 1 CUCCUCUUUGGGUUCU-UUUCUCGGAACGCUCGAGAGUUUCUAUCCCAGUUGUUGCUGGUGUGGGCGUCACAGUUCAAUGUCAGAACUGCCCACACGGGGAUUUAC .(((((...((((...-.(((((((.....))))))).....))))((((....))))((((((((.....(((((.......))))))))))))))))))..... ( -38.90) >DroYak_CAF1 7912 102 - 1 UUCU-UUAUGGGUUCUUUUUCUCGGAAUACCCGAGAGUUUCUACUUAAGUU---GCUGGCGUGGGCGUCACAGUUCAAUGUCAGAACUGCCCACAUGGGGAUUUAC .(((-((((((((.....(((((((.....)))))))..........((((---.(((((((((((......)))).))))))))))))))))..))))))..... ( -33.70) >consensus CUCUUCUUUGGGUUCU_UUUCUCGGAACGCUCGAGAGUUUCUAUCCCAGUUGUUGCUGGUGUGGGCGUCACAGUUCAAUGUCAGAACUGCCCACACGGGGAUUUAC .(((((...((((.....(((((((.....))))))).....))))............((((((((.....(((((.......))))))))))))))))))..... (-32.04 = -31.48 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:04 2006