
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,805,638 – 11,805,730 |
| Length | 92 |
| Max. P | 0.987962 |

| Location | 11,805,638 – 11,805,730 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -21.37 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11805638 92 - 23771897 UUCCUGGAAGUGGUUCAGUACCAGAUGGACAUUCUGGAAUAUUUCCGGGAGAGCGAGGUAAGCUAAACCAGGG----------ACAUAAACAU----AGGCUUCAA .(((((((((((.(((((..((....)).....))))).)))))))))))((((..(((.......)))..(.----------.......)..----..))))... ( -25.40) >DroSec_CAF1 3986 102 - 1 UUCCUGGAAGUGGUUCAGUACCAGAUGGACAUUCUGGAAUACUUCCGGGAGAGCGAGGUAAGCUAAACCGGGGCAUAAGCUAAACAUAAACAU----AGUCUUCAC .(((((((((((.(((((..((....)).....))))).)))))))))))(((.(((((.......)))(.(((....)))...)........----..))))).. ( -29.10) >DroSim_CAF1 4962 85 - 1 UUCCUGGAAGUGGUUCAGUACCAGAUGGACAUUCUGGAAUACUUCCGGGAGAGCGAGGUAAGC-----------------UAAACAUAAACAU----AGUCUUCAC ((((((((((((.(((((..((....)).....))))).))))))))))))...((((...((-----------------((..........)----))))))).. ( -26.90) >DroEre_CAF1 7576 90 - 1 UUCCUGGAAGUGGUCCAGUACCAGAUGGACAUUUUGAAAUACUUCCGGGAAAGCGAGGUAUGU----CCGGGG----------ACAUCAACUCUAA--AACUGCAC (((((((((((((((((........))))).........))))))))))))...(((..((((----(....)----------))))...)))...--........ ( -31.70) >DroYak_CAF1 9806 92 - 1 UUCCUGGAAGUGGUUCAGUACCAGAUGGACAUCCUGGAAUACUUUCGGGAGAGCGAGGUAAGU----CCGGGG----------ACAUCUACUUGCACUAGCUGCAC ((((((((((((.(((((..((....)).....))))).))))))))))))(((.((((((((----..(...----------.)....)))))).)).))).... ( -31.60) >DroAna_CAF1 3833 87 - 1 UUCUUCGAGGUGGUCCAGUACCAGAUGGACAUCCUGGAUAACUUCCGGGAGAGUGAGGUGAGU----UAGUGG-UUA------AGAUUAGC-CUAA--GAU----- ..(((((.....(((((........))))).(((((((.....)))))))...))))).....----....((-(((------....))))-)...--...----- ( -24.90) >consensus UUCCUGGAAGUGGUUCAGUACCAGAUGGACAUUCUGGAAUACUUCCGGGAGAGCGAGGUAAGC____CCGGGG__________ACAUAAACAU____AGACUUCAC ((((((((((((.(((((..((....)).....))))).))))))))))))....................................................... (-21.37 = -20.93 + -0.44)


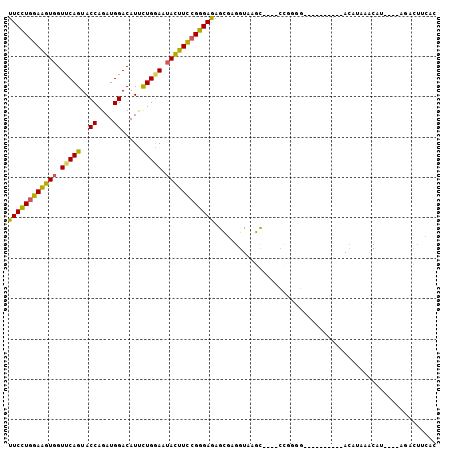
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:26 2006