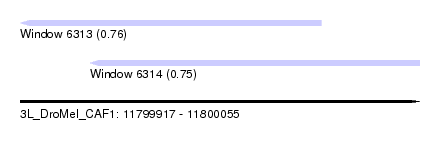
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,799,917 – 11,800,055 |
| Length | 138 |
| Max. P | 0.759011 |

| Location | 11,799,917 – 11,800,021 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.83 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -19.84 |
| Energy contribution | -22.19 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799917 104 - 23771897 CUGAGGGCGACUUUUCGCUUCUUGCAGAGCACGGUGUAUAAGAAAGAGUUGGAUUCGGCCUUGCUGGGUUUGGAACAACAAUAUAUACUGAGAUUAGGAAAAAG (((.((((((....))))))....)))....(((((((((........(..((((((((...))))))))..)........))))))))).............. ( -26.69) >DroSec_CAF1 9453 80 - 1 CUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAUCCUUCCU------------------------GAGCUCGGAUAGGAG .((((..(((((((((.((((((((...)))))).)).....)))))))))..))))((((..((------------------------(....)))..)))). ( -27.50) >DroSim_CAF1 9045 104 - 1 CUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAGCCUUCCUGGAUUUGCAACACCAAUAUUUACUGAGCUCGACUAGGAG .(((.(((((....))))))))....((((..((((((((((((((.((((....))))))))))((..........))..))))))))..))))......... ( -34.30) >consensus CUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAGCCUUCCUGG_UUUG_AACA_CAAUAU_UACUGAGCUCGGAUAGGAG ..((.(((((....))))))).....((((.(((((((((((((((.((((....))))))))))................))))))))).))))......... (-19.84 = -22.19 + 2.35)


| Location | 11,799,941 – 11,800,055 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -29.55 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799941 114 - 23771897 ------CCAUAUAAGCCCCGCUACGUCGUUCCAGGAAUGCCUGAGGGCGACUUUUCGCUUCUUGCAGAGCACGGUGUAUAAGAAAGAGUUGGAUUCGGCCUUGCUGGGUUUGGAACAACA ------...((((.(((..(((..(((((((((((....)))).))))))).....((.....))..)))..))).)))).......((((..((..((((....))))..))..)))). ( -35.50) >DroSec_CAF1 9468 105 - 1 CACAGACCCUAUCAGCCCCACUAUGUCGUUCCAGGAAUGGCUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAUCCUUCCU--------------- ......((((((.....((((((((((((((...)))))(((((.(((((....)))))))).))...))))))))))))))).......(((........))).--------------- ( -36.00) >DroSim_CAF1 9069 120 - 1 CACAGACCCUAUCAGCCCCACCAUGUCGUUCCAGGAAUGGCUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAGCCUUCCUGGAUUUGCAACACCA .......................(((...((((((((.(((((((..(((((((((.((((((((...)))))).)).....)))))))))..)))))))))))))))...)))...... ( -46.00) >consensus CACAGACCCUAUCAGCCCCACUAUGUCGUUCCAGGAAUGGCUGAAGGCGACUUUUCGCCUCAUGCAGAGCGUGGUGGAUAGGGAAGAGUUGGAUUCAGCCUUCCUGG_UUUG_AACA_CA .................((((((((((((((...))))).(((.((((((....))))))....))).)))))))))...((((((.((((....))))))))))............... (-29.55 = -29.90 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:25 2006