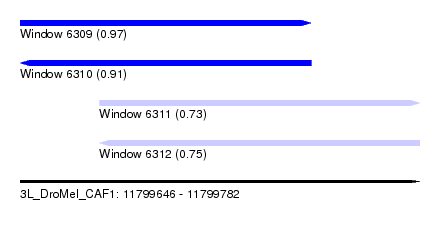
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,799,646 – 11,799,782 |
| Length | 136 |
| Max. P | 0.967683 |

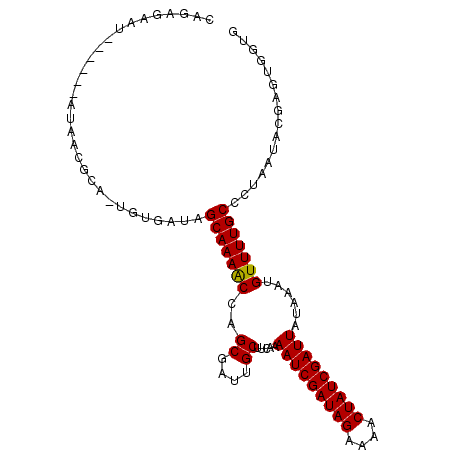
| Location | 11,799,646 – 11,799,745 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799646 99 + 23771897 CACCAAUCGUAUAAGGGCAAAACAUUUAUAAUCGAUAGUUUUCUAUCGAUUAUAAAGCAAUCGCGGGCUUUGCAUUCAC----UGUUAUUCAGGCAUUCAUUG ........((......(((((.(.((((((((((((((....))))))))))))))((....))..).))))).....(----((.....)))))........ ( -24.30) >DroSec_CAF1 9182 97 + 1 CAACACUCGUAUUAGGGCAAAACAUUUAUAAUCGAUAGUUGUCUAUCGAUUUUGAAGCAAUCGCUGGUUUUGCUAUCGCAUUGCGUUAU------AUCCUCUG .............(((..(.(((.((((.(((((((((....))))))))).))))(((((((.(((.....))).)).)))))))).)------..)))... ( -25.50) >DroSim_CAF1 8777 97 + 1 CACCACUCGUAUUAGGGCAAAACAUUUAUAAUCGAUAGUUUUCUAUCGAUUUUCAAGCAAUCGCUGGUUUUGCUAUCACACUGCGUUAU------AUUCUCUG .......((((....((((((((......(((((((((....)))))))))....(((....))).)))))))).......))))....------........ ( -23.10) >consensus CACCACUCGUAUUAGGGCAAAACAUUUAUAAUCGAUAGUUUUCUAUCGAUUUUAAAGCAAUCGCUGGUUUUGCUAUCACA_UGCGUUAU______AUUCUCUG ................(((((((.((((.(((((((((....))))))))).))))((....))..))))))).............................. (-19.18 = -19.73 + 0.56)

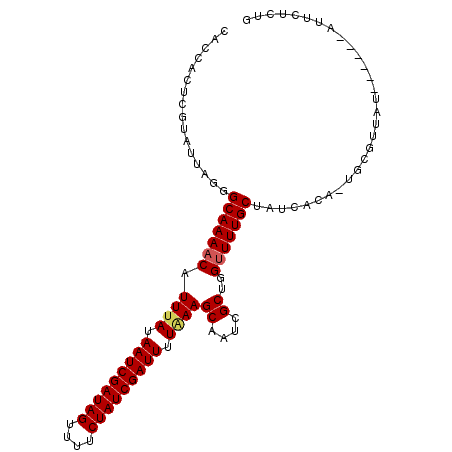

| Location | 11,799,646 – 11,799,745 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799646 99 - 23771897 CAAUGAAUGCCUGAAUAACA----GUGAAUGCAAAGCCCGCGAUUGCUUUAUAAUCGAUAGAAAACUAUCGAUUAUAAAUGUUUUGCCCUUAUACGAUUGGUG ........(((..(((....----((((..(((((((..((....))((((((((((((((....)))))))))))))).)))))))..))))...)))))). ( -25.60) >DroSec_CAF1 9182 97 - 1 CAGAGGAU------AUAACGCAAUGCGAUAGCAAAACCAGCGAUUGCUUCAAAAUCGAUAGACAACUAUCGAUUAUAAAUGUUUUGCCCUAAUACGAGUGUUG ...(((..------....(((...)))...(((((((.(((....)))....(((((((((....)))))))))......))))))))))............. ( -22.70) >DroSim_CAF1 8777 97 - 1 CAGAGAAU------AUAACGCAGUGUGAUAGCAAAACCAGCGAUUGCUUGAAAAUCGAUAGAAAACUAUCGAUUAUAAAUGUUUUGCCCUAAUACGAGUGGUG ........------....(((.((((....(((((((.(((....)))....(((((((((....)))))))))......)))))))....))))..)))... ( -24.60) >consensus CAGAGAAU______AUAACGCA_UGUGAUAGCAAAACCAGCGAUUGCUUCAAAAUCGAUAGAAAACUAUCGAUUAUAAAUGUUUUGCCCUAAUACGAGUGGUG ..............................(((((((..((....)).....(((((((((....)))))))))......)))))))................ (-17.75 = -17.53 + -0.22)


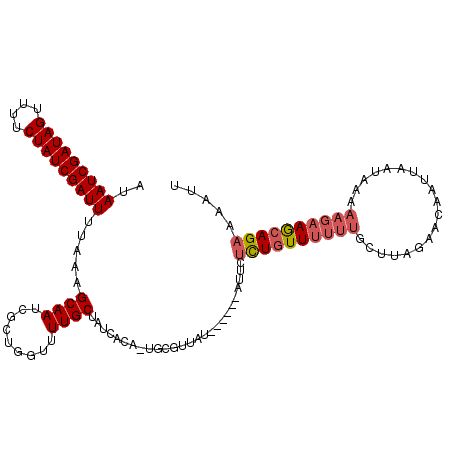
| Location | 11,799,673 – 11,799,782 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -13.98 |
| Energy contribution | -15.54 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799673 109 + 23771897 AUAAUCGAUAGUUUUCUAUCGAUUAUAAAGCAAUCGCGGGCUUUGCAUUCAC----UGUUAUUCAGGCAUUCAUUGUUA---UCCUAAAACAGUUAAUAAAAAGGAAAAGAAAAAU (((((((((((....)))))))))))...((....))...((((..(((.((----((((....(((.((........)---))))..)))))).)))..))))............ ( -22.80) >DroSec_CAF1 9209 110 + 1 AUAAUCGAUAGUUGUCUAUCGAUUUUGAAGCAAUCGCUGGUUUUGCUAUCGCAUUGCGUUAU------AUCCUCUGUUUUUUGCUUAGAAUAAUUAAUAAAAAGAAGCAGAAAAUU ..(((((((((....))))))))).....(((((((.(((.....))).)).))))).....------....((((((((((..(((..........))).))))))))))..... ( -26.90) >DroSim_CAF1 8804 110 + 1 AUAAUCGAUAGUUUUCUAUCGAUUUUCAAGCAAUCGCUGGUUUUGCUAUCACACUGCGUUAU------AUUCUCUGUUUUUUGCUUAGAACAAUUAAUAAAAAGAAGCAGAAAAUU ..(((((((((....)))))))))....(((((.........)))))......((((.((.(------(((...((((((......))))))...)))).))....))))...... ( -22.10) >consensus AUAAUCGAUAGUUUUCUAUCGAUUUUAAAGCAAUCGCUGGUUUUGCUAUCACA_UGCGUUAU______AUUCUCUGUUUUUUGCUUAGAACAAUUAAUAAAAAGAAGCAGAAAAUU ..(((((((((....))))))))).....((((.........))))..........................((((((((((...................))))))))))..... (-13.98 = -15.54 + 1.56)


| Location | 11,799,673 – 11,799,782 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11799673 109 - 23771897 AUUUUUCUUUUCCUUUUUAUUAACUGUUUUAGGA---UAACAAUGAAUGCCUGAAUAACA----GUGAAUGCAAAGCCCGCGAUUGCUUUAUAAUCGAUAGAAAACUAUCGAUUAU ............((((.((((.((((((((((((---(........)).)))))..))))----)).)))).))))...((....))...(((((((((((....))))))))))) ( -23.90) >DroSec_CAF1 9209 110 - 1 AAUUUUCUGCUUCUUUUUAUUAAUUAUUCUAAGCAAAAAACAGAGGAU------AUAACGCAAUGCGAUAGCAAAACCAGCGAUUGCUUCAAAAUCGAUAGACAACUAUCGAUUAU .((((((((....(((((.(((.......)))..))))).))))))))------..........(((((.((.......)).))))).....(((((((((....))))))))).. ( -20.90) >DroSim_CAF1 8804 110 - 1 AAUUUUCUGCUUCUUUUUAUUAAUUGUUCUAAGCAAAAAACAGAGAAU------AUAACGCAGUGUGAUAGCAAAACCAGCGAUUGCUUGAAAAUCGAUAGAAAACUAUCGAUUAU .((((((((....(((((.(((.......)))..))))).))))))))------......(((.(..((.((.......)).))..))))..(((((((((....))))))))).. ( -20.20) >consensus AAUUUUCUGCUUCUUUUUAUUAAUUGUUCUAAGCAAAAAACAGAGAAU______AUAACGCA_UGUGAUAGCAAAACCAGCGAUUGCUUCAAAAUCGAUAGAAAACUAUCGAUUAU ................................................................(((((.((.......)).))))).....(((((((((....))))))))).. (-13.75 = -13.87 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:23 2006