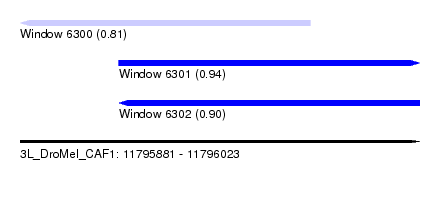
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,795,881 – 11,796,023 |
| Length | 142 |
| Max. P | 0.943557 |

| Location | 11,795,881 – 11,795,984 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11795881 103 - 23771897 UUAAAACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAGUGUUUAGUCGGCUUUUCGCUUUCGUUAGAUAAAAA ....(((((((((((.((((((..((.....))..)))))))).)))(((((((((.(((.....)))...)))))))))........))))))......... ( -28.00) >DroSec_CAF1 5182 101 - 1 CCAACACAGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG--UUUAGUCGGCUUUUCGCUUUCGUUAGAUAAAAA ........(((((((.((((((..((.....))..))))))......(((((((((.(((.....)))--.)))))))))....)))))))............ ( -27.00) >DroSim_CAF1 4790 101 - 1 CCAACACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG--UUUAGUCGGCUUUUCGCUUUCGUUAGAUAAAAA .....((((((((((.((((((..((.....))..)))))))).)))(((((((((.(((.....)))--.)))))))))........))))).......... ( -27.40) >consensus CCAACACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG__UUUAGUCGGCUUUUCGCUUUCGUUAGAUAAAAA ........(((((((.((((((..((.....))..))))))......(((((((((.(((.....)))...)))))))))....)))))))............ (-25.50 = -25.50 + 0.00)


| Location | 11,795,916 – 11,796,023 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 97.51 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -34.04 |
| Energy contribution | -34.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11795916 107 + 23771897 CUGAUAUGCAGACUAAUCGGCGAGAUGGUGAAAGCAAUGUUUUCAUUUCCACGCACUCUCCGUUUUAAGGAGAGAGCGCAAGAGAGUUUCUCUUUGCCUCUCUUGAA .......((..........(((.(..(((((((((...)))))))))..).))).((((((.......)))))).)).((((((((...........)))))))).. ( -34.10) >DroSec_CAF1 5215 107 + 1 CUGAUAUGCAGACUAAUCGGCGAGAUGGUGAAAGCAAUGUUUUCAUUUCCACGCACUCUCUGUGUUGGGGAGAGAGCGCAAGAGAGUUUCUCUUUGCCUCUCUUGAA (..((..(((((.......(((.(..(((((((((...)))))))))..).)))....)))))))..).(((((((.(((((((.....))).)))))))))))... ( -36.00) >DroSim_CAF1 4823 107 + 1 CUGAUAUGCAGACUAAUCGGCGAGAUGGUGAAAGCAAUGUUUUCAUUUCCACGCACUCUCCGUGUUGGGGAGAGAGCGCAAGAGAGUUUCUCUUUGCCUCUCUUGAA (..(((((.(((.......(((.(..(((((((((...)))))))))..).)))..))).)))))..).(((((((.(((((((.....))).)))))))))))... ( -37.10) >consensus CUGAUAUGCAGACUAAUCGGCGAGAUGGUGAAAGCAAUGUUUUCAUUUCCACGCACUCUCCGUGUUGGGGAGAGAGCGCAAGAGAGUUUCUCUUUGCCUCUCUUGAA ((((((((.(((.......(((.(..(((((((((...)))))))))..).)))..))).)))))))).(((((((.(((((((.....))).)))))))))))... (-34.04 = -34.27 + 0.23)


| Location | 11,795,916 – 11,796,023 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 97.51 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11795916 107 - 23771897 UUCAAGAGAGGCAAAGAGAAACUCUCUUGCGCUCUCUCCUUAAAACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG .....((((.((((.(((.....)))))))((.((((((.......)))))).))((((((..((.....))..))))))..))))((.((....)).))....... ( -31.70) >DroSec_CAF1 5215 107 - 1 UUCAAGAGAGGCAAAGAGAAACUCUCUUGCGCUCUCUCCCCAACACAGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG ....((((((((((.(((.....))))))).))))))..........((((.((.((((((..((.....))..))))))))))))((.((....)).))....... ( -29.20) >DroSim_CAF1 4823 107 - 1 UUCAAGAGAGGCAAAGAGAAACUCUCUUGCGCUCUCUCCCCAACACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG .....((((.((((.(((.....)))))))((.((((((.......)))))).))((((((..((.....))..))))))..))))((.((....)).))....... ( -31.20) >consensus UUCAAGAGAGGCAAAGAGAAACUCUCUUGCGCUCUCUCCCCAACACGGAGAGUGCGUGGAAAUGAAAACAUUGCUUUCACCAUCUCGCCGAUUAGUCUGCAUAUCAG .....((((.((((.(((.....)))))))((.((((((.......)))))).))((((((..((.....))..))))))..))))((.((....)).))....... (-29.63 = -29.97 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:13 2006