
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,794,449 – 11,794,809 |
| Length | 360 |
| Max. P | 0.925988 |

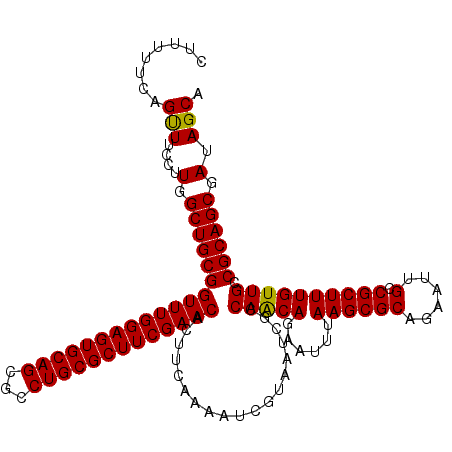
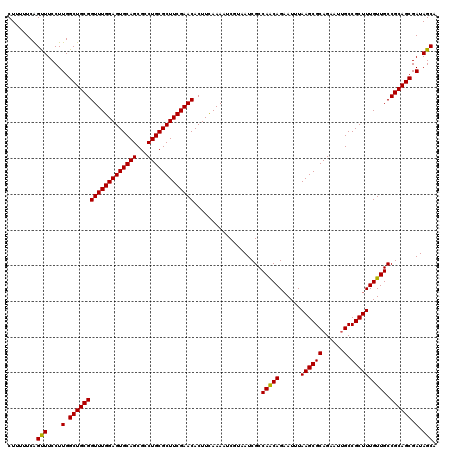
| Location | 11,794,449 – 11,794,569 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -39.77 |
| Energy contribution | -39.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794449 120 + 23771897 CUUUUUCGGUUUCCUUGGCUGCGGUUUGGAGUGCAGCGCCUGCGCUUCGAACACUUUAAAAUCAUAAUCGCCAGCAGCAUUUAAGCGCAGAAUAGCCGCUUUGUUGCCGCAGCGAUAGCA ........(((....(.(((((((((((((((((((...))))))))))))).....................((((((...((((((......).))))))))))))))))).).))). ( -43.10) >DroSec_CAF1 3470 120 + 1 CUUUUUCAGUUUCCUUGGCUGCGGUUUGGAGUGCAGCGCCUGCGCUUCGAACACUUCAAAAUCGUAAUCGCCAACAGAAUUUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCA ........(((....(.(((((((((((((((((((...)))))))))))))...................(((((......((((((......).)))))))))).)))))).).))). ( -38.70) >DroSim_CAF1 3347 120 + 1 CUUUUUCAGCUUCCUUGGCUGCGGUUUGGAGUGCAGCGCCUGCGCUUCGAACACUUCAAAAUCGUAAUCGCCAACAGAAUUUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCA ........(((....(.(((((((((((((((((((...)))))))))))))...................(((((......((((((......).)))))))))).)))))).).))). ( -41.10) >consensus CUUUUUCAGUUUCCUUGGCUGCGGUUUGGAGUGCAGCGCCUGCGCUUCGAACACUUCAAAAUCGUAAUCGCCAACAGAAUUUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCA ........(((....(.(((((((((((((((((((...)))))))))))))...................(((((......((((((......).)))))))))).)))))).).))). (-39.77 = -39.33 + -0.44)

| Location | 11,794,449 – 11,794,569 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -36.31 |
| Energy contribution | -36.09 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794449 120 - 23771897 UGCUAUCGCUGCGGCAACAAAGCGGCUAUUCUGCGCUUAAAUGCUGCUGGCGAUUAUGAUUUUAAAGUGUUCGAAGCGCAGGCGCUGCACUCCAAACCGCAGCCAAGGAAACCGAAAAAG .....(((((((((.......(((((...(((((((((.((((((....................))))))..))))))))).)))))........)))))))...(....).))..... ( -38.01) >DroSec_CAF1 3470 120 - 1 UGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAAAUUCUGUUGGCGAUUACGAUUUUGAAGUGUUCGAAGCGCAGGCGCUGCACUCCAAACCGCAGCCAAGGAAACUGAAAAAG .....(((((((((.......(((((...(((((((((...(((.((((.......))))...))).......))))))))).)))))........)))))))..((....))))..... ( -39.86) >DroSim_CAF1 3347 120 - 1 UGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAAAUUCUGUUGGCGAUUACGAUUUUGAAGUGUUCGAAGCGCAGGCGCUGCACUCCAAACCGCAGCCAAGGAAGCUGAAAAAG .(((.(((((((((.......(((((...(((((((((...(((.((((.......))))...))).......))))))))).)))))........)))))))...)).)))........ ( -39.56) >consensus UGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAAAUUCUGUUGGCGAUUACGAUUUUGAAGUGUUCGAAGCGCAGGCGCUGCACUCCAAACCGCAGCCAAGGAAACUGAAAAAG .....(((((((((.......(((((...(((((((((.....(....).(((.(((.........))).)))))))))))).)))))........)))))))...(....).))..... (-36.31 = -36.09 + -0.22)



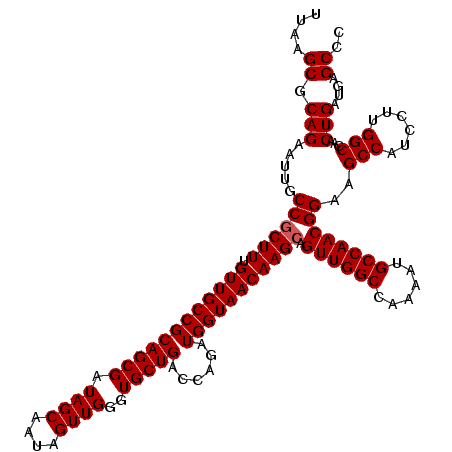
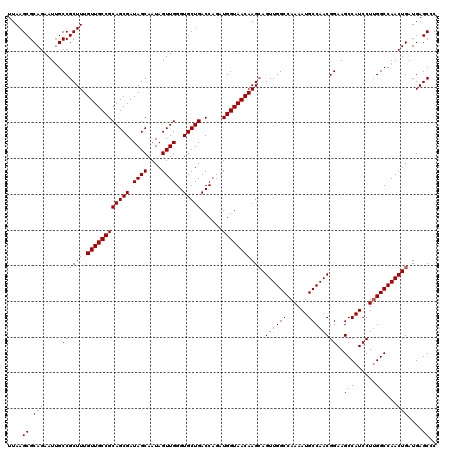
| Location | 11,794,529 – 11,794,649 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -37.87 |
| Energy contribution | -38.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794529 120 + 23771897 UUAAGCGCAGAAUAGCCGCUUUGUUGCCGCAGCGAUAGCAAUAGUUGGGUGCUGACCAUAUGGUAACAAGAAGUUGGCCAAAAUGCCAACGGAAGCCAUCCUGGGCCAACUGAUGAGCGC ....((((...........(((((((((((((((.((((....))))..)))))......)))))))))).((((((((...(((....(....).)))....)))))))).....)))) ( -39.20) >DroSec_CAF1 3550 120 + 1 UUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCAAUAGUUGGGUGCUGACCAGAUGGUAACAAGCAGUUGGCCAAAAUGCCAACGGAAGCCAUCCUUGGCCAACUGAUGAGCCC ....((.(((.....((((((.((((((((((((.((((....))))..)))))......))))))))))).((((((......))))))))..((((....))))...)))....)).. ( -41.20) >DroSim_CAF1 3427 120 + 1 UUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCAAUAGUUGGGUGCUGACCAGAUGGUAACAAGCAGUUGGCCAAAAUGCCAACGGAAGCCAUCCUUGGCCAACUGAUGAGCCC ....((.(((.....((((((.((((((((((((.((((....))))..)))))......))))))))))).((((((......))))))))..((((....))))...)))....)).. ( -41.20) >consensus UUAAGCGCAGAAUUGCCGCUUUGUUGCCGCAGCGAUAGCAAUAGUUGGGUGCUGACCAGAUGGUAACAAGCAGUUGGCCAAAAUGCCAACGGAAGCCAUCCUUGGCCAACUGAUGAGCCC ....((.(((.....((((((.((((((((((((.((((....))))..)))))......))))))))))).((((((......))))))))..(((......)))...)))....)).. (-37.87 = -38.20 + 0.33)

| Location | 11,794,529 – 11,794,649 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -37.67 |
| Energy contribution | -38.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794529 120 - 23771897 GCGCUCAUCAGUUGGCCCAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUUCUUGUUACCAUAUGGUCAGCACCCAACUAUUGCUAUCGCUGCGGCAACAAAGCGGCUAUUCUGCGCUUAA ((((........(((((...((((((....((((((......))))))....(((((((....))).))))..........))))))(((..(....)..))))))))....)))).... ( -40.50) >DroSec_CAF1 3550 120 - 1 GGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAUUGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAA (((.....(((((((((((((...(((......))).)))))))))))))..(((((((....))).))))))).......((...((((..(....)..))))(((....))))).... ( -42.70) >DroSim_CAF1 3427 120 - 1 GGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAUUGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAA (((.....(((((((((((((...(((......))).)))))))))))))..(((((((....))).))))))).......((...((((..(....)..))))(((....))))).... ( -42.70) >consensus GGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAUUGCUAUCGCUGCGGCAACAAAGCGGCAAUUCUGCGCUUAA (((.....(((((((((.((((((.(.......).)))))))))))))))..(((((((....))).))))))).......((...((((..(....)..))))((......)))).... (-37.67 = -38.33 + 0.67)



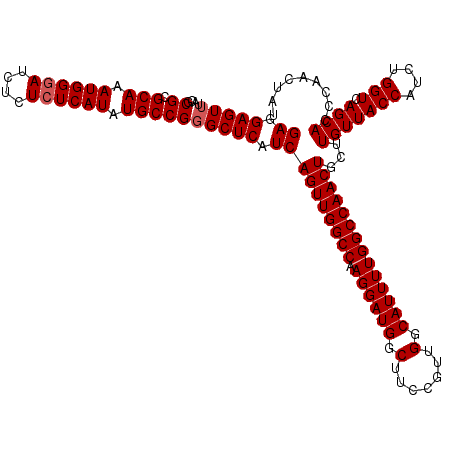
| Location | 11,794,569 – 11,794,689 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -37.97 |
| Energy contribution | -38.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794569 120 - 23771897 GAGGAGUUUACGCGCGCAAAUGGAAUCUCUCUCAUAUGCCGCGCUCAUCAGUUGGCCCAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUUCUUGUUACCAUAUGGUCAGCACCCAACUAU ((.(((....((((.(((.(((((.....)).))).)))))))))).))((((((((.((((((.(.......).))))))))))))))...(((((((....))).))))......... ( -39.30) >DroSec_CAF1 3590 120 - 1 GAGGAGUUUACGCGCGCAAAUGGGAUCUCUCUCAUAUGCCGGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAU ....((((.....(.(((.((((((....)))))).))))(((.....(((((((((((((...(((......))).)))))))))))))..(((((((....))).))))))))))).. ( -42.40) >DroSim_CAF1 3467 120 - 1 GAGGAGUUUACGCGCGCAAAUGGGAUCUCUCUCAUAUGCCGGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAU ....((((.....(.(((.((((((....)))))).))))(((.....(((((((((((((...(((......))).)))))))))))))..(((((((....))).))))))))))).. ( -42.40) >consensus GAGGAGUUUACGCGCGCAAAUGGGAUCUCUCUCAUAUGCCGGGCUCAUCAGUUGGCCAAGGAUGGCUUCCGUUGGCAUUUUGGCCAACUGCUUGUUACCAUCUGGUCAGCACCCAACUAU ((.(((((....((.(((.((((((....)))))).)))))))))).))((((((((.((((((.(.......).))))))))))))))...(((((((....))).))))......... (-37.97 = -38.63 + 0.67)


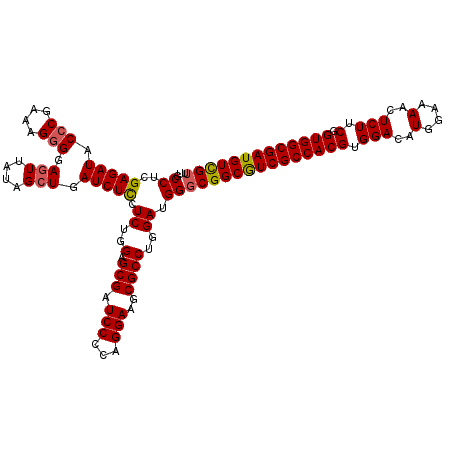
| Location | 11,794,689 – 11,794,809 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -48.13 |
| Consensus MFE | -41.63 |
| Energy contribution | -41.97 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11794689 120 - 23771897 UGCCACGAGAUACCCGAAAGUGGAAUUAUAGCUGAUCUUCUCUGGAGCGAUCCACAGGAAGCGCCUGGAUGGGCGGCAUCGCCACGAGGACAUGGAAAACUCUUCGGUGGCGAUGUUGUU ..((((.((...(((....).)).......(((....((((.((((....)))).)))))))..)).).)))((((((((((((((((((..(....)..))))).))))))))))))). ( -44.70) >DroSec_CAF1 3710 120 - 1 UGCCUCGAGAUACCCGAAAGGGGAGUUAUAGCUGAUCUCCUCUGGAGCGAUCCCCAGGAAGCGCCUGGAUGGGCGGCGUCGCCACGUGGACAUGGAAAACUCUUCGGUGGCGAUGUCGUU .((((.(((((.(((....))).(((....))).))))).((..(.(((.(((...)))..))))..)).))))(((((((((((.((((...(.....)..)))))))))))))))... ( -50.30) >DroSim_CAF1 3587 120 - 1 UUCCUCGAGAUACCCGAAAGGGGAGUUAUAGCUGAUCUCCUCUGGAGCGAUCCCCAGGAAGCGCCUGGAUGGUCGGCGUCGCCACGUGGACAUGGAAAACUCUUCGGUGGCGAUGUCGUU ((((..(((((.(((....))).(((....))).)))))....)))).((((.(((((.....)))))..))))(((((((((((.((((...(.....)..)))))))))))))))... ( -49.40) >consensus UGCCUCGAGAUACCCGAAAGGGGAGUUAUAGCUGAUCUCCUCUGGAGCGAUCCCCAGGAAGCGCCUGGAUGGGCGGCGUCGCCACGUGGACAUGGAAAACUCUUCGGUGGCGAUGUCGUU ..((..(((((.(((....))).(((....))).))))).((..(.(((.(((...)))..))))..)).))((((((((((((((.(((..(....)..))).).))))))))))))). (-41.63 = -41.97 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:11 2006