| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,775,404 – 11,775,513 |
| Length | 109 |
| Max. P | 0.947760 |

| Location | 11,775,404 – 11,775,513 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -23.98 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
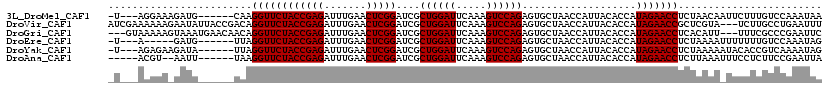
>3L_DroMel_CAF1 11775404 109 + 23771897 -U---AGGAAAGAUG------CAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCUAACAAUUCUUUGUCCAAAUAA -.---.(((((((((------..(((((((((((((.......)))))....((((((.....))))))...................))))))))....))..))))..)))...... ( -28.40) >DroVir_CAF1 11245 116 + 1 AUCGAAAAAAGAAUAUUACCGACAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCUCGUA---UCUUGCCUGAAUUU .(((....((((....(((.((..((((((((((((.......)))))....((((((.....))))))...................)))))))..)))))---))))...))).... ( -27.50) >DroGri_CAF1 11330 113 + 1 ---GUAAAAAGUAAAUGAACAACAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCACAUU---UUUCGCCCGAAUUC ---(..((((((...(((......((((((((((((.......)))))....((((((.....))))))...................)))))))))).)))---)))..)........ ( -25.00) >DroEre_CAF1 11264 104 + 1 -U---A-----GAUG------UUAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCUAAAAUUUUUUUGUCCAAAUAG -.---.-----....------..(((((((((((((.......)))))....((((((.....))))))...................))))))))....................... ( -24.40) >DroYak_CAF1 11338 109 + 1 -U---AGAGAAGAUA------UUAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCUAAAAAUACACCGUCAAAAUAG -.---..........------..(((((((((((((.......)))))....((((((.....))))))...................))))))))....................... ( -24.40) >DroAna_CAF1 10765 106 + 1 -----ACGU--AAUU------UAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCUUAAAUUUCCUCUUCCGAAUUA -----..(.--((((------(((((((((((((((.......)))))....((((((.....))))))...................))))))))..)))))).)............. ( -25.90) >consensus _U___AAAAA_GAUA______UAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCUAAAA_UUCUUCGUCCAAAUAA ........................((((((((((((.......)))))....((((((.....))))))...................)))))))........................ (-23.98 = -23.98 + -0.00)

| Location | 11,775,404 – 11,775,513 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11775404 109 - 23771897 UUAUUUGGACAAAGAAUUGUUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUG------CAUCUUUCCU---A- ......(((..((((..(((..(((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))------))))))))).---.- ( -31.10) >DroVir_CAF1 11245 116 - 1 AAAUUCAGGCAAGA---UACGAGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGUCGGUAAUAUUCUUUUUUCGAU ....(((((.((((---..(((..(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..))).......)))).))).)). ( -27.00) >DroGri_CAF1 11330 113 - 1 GAAUUCGGGCGAAA---AAUGUGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGUUGUUCAUUUACUUUUUAC--- (((((((((((...---....((((((((...(((((........)))))..)))))))).......)).)))))))))...........((((.....)))).............--- ( -29.14) >DroEre_CAF1 11264 104 - 1 CUAUUUGGACAAAAAAAUUUUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAA------CAUC-----U---A- .......................((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..------....-----.---.- ( -24.90) >DroYak_CAF1 11338 109 - 1 CUAUUUUGACGGUGUAUUUUUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAA------UAUCUUCUCU---A- .......((.(((((........((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))).)------)))).))...---.- ( -26.40) >DroAna_CAF1 10765 106 - 1 UAAUUCGGAAGAGGAAAUUUAAGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUA------AAUU--ACGU----- ...(((....)))(.((((((..((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))------))))--.)..----- ( -29.40) >consensus CAAUUCGGACAAAAAA_UUUUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAA______UAUC_UUUCU___A_ .......................((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))....................... (-24.00 = -24.17 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:54 2006