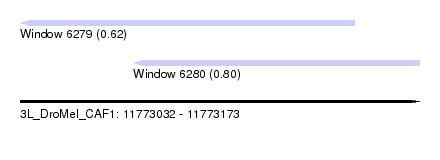
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,773,032 – 11,773,173 |
| Length | 141 |
| Max. P | 0.803673 |

| Location | 11,773,032 – 11,773,150 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -23.34 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11773032 118 - 23771897 AUAUAUUUAACUAAUAU--UAAAUUUCAGCCCGUGGCAUCUAUGUCAUCGGAUUUAGCUAUCCGGUGGUUCCUCAGGGAAAGGCACGCAUCCGUGUUCAAAUCUCCGCUGCCCACACGGA .................--...........(((((.........(((((((((......))))))))).......(((...(((((((....))))..........))).))).))))). ( -29.40) >DroVir_CAF1 8731 115 - 1 UAAAAUUUG-----UUUUAAUUCCUGCAGCACGCGGCAUUUAUGUCAUAGGCUUCAGCUAUCCGGUGGUGCCACAAGGCAAGGCACGCAUCCGCGUCCAAAUCUCAGCGGCUCACACCGA .........-----.........(((.(((....((((....))))....))).))).....((((((.(((.(.(((...((.((((....))))))...)))..).))).).))))). ( -31.00) >DroGri_CAF1 8455 118 - 1 AUACAGUUGCC-A-CUUUAUUUAUUGCAGCACGUGGCAUUUAUGUCAUUGGAUUCAGCUAUCCAGUGGUGCCGCAGGGCAAGGCACGCAUUCGCGUUCAAAUCUCCGCCGCUCACACAGA .....(((((.-.-...........)))))..(((((((......((((((((......))))))))))))))).((((..(((((((....))))..........)))))))....... ( -35.42) >DroSec_CAF1 8743 117 - 1 AUAUAUUAAACU-AAAU--UCAUUUCCAGCCCGUGGCAUCUAUGUCAUCGGAUUUAGCUAUCCGGUGGUUCCGCAAGGAAAGGCACGCAUCCGUGUCCAAAUAUCCGCUGCUCACACGGA ............-....--...........((((((((.(....(((((((((......)))))))))((((....)))).((((((....)))))).........).)))...))))). ( -34.00) >DroYak_CAF1 8931 118 - 1 CUGUAAUAAACCAAUCA--UAAAUUUCAGCCCGUGGCAUCUACGUUAUCGGAUUCAGCUAUCCGGUGGUUCCCCAAGGAAAGGCACGCAUCCGUGUUCAAAUCUCCGCUGCCCACACGGA .................--...........((((((((..........(((((......)))))((((((((....)))).((((((....)))))).......))))))))...)))). ( -27.70) >DroAna_CAF1 8583 117 - 1 CCUUAUUAAAGCUA-GU--ACCUUUACAGCUCGAGGUAUUUAUGUCAUUGGCUUCAGUUACCCAGUGGUUCCCCAGGGCAAGGCCCGCAUCCGCGUUCAAAUCUCUGCCGCCCAUACUGA ........((((((-((--(((((........))))))).........))))))((((......(((((......((((...))))((....))............)))))....)))). ( -26.20) >consensus AUAUAUUAAAC_AAUAU__UAAAUUGCAGCCCGUGGCAUCUAUGUCAUCGGAUUCAGCUAUCCGGUGGUUCCCCAAGGAAAGGCACGCAUCCGCGUUCAAAUCUCCGCUGCCCACACGGA ................................(((((.......(((((((((......)))))))))((((....)))).((.((((....))))))........)))))......... (-23.34 = -22.82 + -0.53)

| Location | 11,773,072 – 11,773,173 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -16.40 |
| Energy contribution | -15.85 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11773072 101 - 23771897 CUUAAUCAUCCCUUU---AUCUACAUAUAUAUUUAACUAAUAUUAAAUUUCAGCCCGUGGCAUCUAUGUCAUCGGAUUUAGCUAUCCGGUGGUUCCUCAGGGAA ........(((((..---....(((((...((((((......))))))....(((...)))...)))))((((((((......)))))))).......))))). ( -22.60) >DroPse_CAF1 8956 96 - 1 CCUAAGGAUCUCUUUUCCAUCCACU----CAUUG---CAA-UUUCUGUCUUAGCUCGCGGCAUCUACGUGAUUGGAUUCAGUUAUCCGGUGGUGCCCCAGGGCA ((...((((..((..((((..(((.----...((---(..-.....((....)).....))).....)))..))))...))..))))...))((((....)))) ( -23.80) >DroSec_CAF1 8783 100 - 1 CUUAAUCAUACCUUU---AUAUACAUAUAUAUUAAACU-AAAUUCAUUUCCAGCCCGUGGCAUCUAUGUCAUCGGAUUUAGCUAUCCGGUGGUUCCGCAAGGAA ......((((....(---((((....))))).......-.............(((...)))...))))(((((((((......)))))))))((((....)))) ( -20.90) >DroSim_CAF1 8017 101 - 1 CUUAAUUAUACCUUU---AUCUACAUAUAUAUUAAACUUAUAUUAAAUUUCAGCCCGUGGCAUCUAUGUCAUCGGAUUUAGCUAUCCGGUGGUUCCUCAGGGAA ..........((((.---....(((((((((.......))))..........(((...)))...)))))((((((((......)))))))).......)))).. ( -18.20) >DroEre_CAF1 9000 100 - 1 CUUAACUAUCGCCUU---AUGUAU-UCCACAUUAAACUAAUCAUAAAUUUCAGCCCGUGGCAUCUACGUUAUCGGAUUUAGCUAUCCGGUGGUUCCUCAAGGAA ...........((((---..(...-.((((......................(((...)))...........(((((......)))))))))...)..)))).. ( -18.00) >DroYak_CAF1 8971 100 - 1 CUGAAUAAUUCCUUU---AUCUAU-UCUGUAAUAAACCAAUCAUAAAUUUCAGCCCGUGGCAUCUACGUUAUCGGAUUCAGCUAUCCGGUGGUUCCCCAAGGAA ........((((((.---...(((-(....))))((((..............(((...))).........(((((((......)))))))))))....)))))) ( -17.00) >consensus CUUAAUCAUCCCUUU___AUCUACAUAUAUAUUAAACUAAUAUUAAAUUUCAGCCCGUGGCAUCUACGUCAUCGGAUUUAGCUAUCCGGUGGUUCCUCAAGGAA ....................................................(((...))).......(((((((((......)))))))))((((....)))) (-16.40 = -15.85 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:51 2006