| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,746,956 – 11,747,070 |
| Length | 114 |
| Max. P | 0.540087 |

| Location | 11,746,956 – 11,747,070 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11746956 114 + 23771897 UAUCGUAACUUGUGUGGCUUCGAACUUACCUAGUUGAG----GAAA-ACUCCCAUGU-CGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUUCGCAUUUUAUCUU ....((((..(((((((((.((((..((((.....(((----(.((-(.(((.....-.))).)))....)))).)))))))))))))...((((((....))))))))))..))))... ( -30.90) >DroVir_CAF1 68970 115 + 1 UAUCGUAACUUGUGUGGCUU-GAAUGUACCUGGUUGAG----GAACGAAUUCAACGUUUGGAUUUUGUUGCCUUUGGUAUUCGAGCCAAUAAGUUCGGCCACCGGUUCACAUUUUAUCUU ..(((.(((((((.((((((-((..(((((.....(((----(((((((.((........)).)))))).)))).)))))))))))))))))))))))...................... ( -34.50) >DroGri_CAF1 57886 100 + 1 UAUCGUAACUUGUGUGGCUUCGAAUGUACCUGGCUAAG----GAAA---------GU-UGGAUUU------CCUAGGUAUUCGAGCCAAUAAGUUCGGCAGCGGGUUCGCAUUUUAUCUU ..(((.(((((((.(((((.(((..((((((.....((----((((---------..-....)))------)))))))))))))))))))))))))))..(((....))).......... ( -32.80) >DroMoj_CAF1 73094 119 + 1 UAUCGUAACUUGUGUGGCUUCGAAUGUACCUCGUCGAGCGAAAAGCGAAUUCAUUGU-UGGAUUUUGUUGCUCUUGGUAUUCGAGCCAAUAAGUUCGGCAACCAGUUCGCAUUUUAUCUU ..(((.(((((((.(((((.(((..(((((.....((((((.....(((((((....-)))))))..))))))..)))))))))))))))))))))))...................... ( -34.40) >DroAna_CAF1 49719 112 + 1 UAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAG----GAAA-AAUC--AGUU-UGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUUCGCAUUUUAUCUU ....((((..(((((((((.(((..(((((.....(((----(.((-((((--....-..))))))....)))).)))))))))))))...((((((....))))))))))..))))... ( -32.30) >DroPer_CAF1 57537 113 + 1 UAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAG----GAAA-ACUCCGA-AA-UGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAAUCGCAUUUUAUCUU ..(((.(((((((.(((((.(((..(((((.....(((----(.((-(.(((..-..-.))).)))....)))).)))))))))))))))))))))))...................... ( -30.70) >consensus UAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAG____GAAA_AAUCC_A_GU_UGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUUCGCAUUUUAUCUU ..(((.(((((((.(((((.(((..(((((.............................................)))))))))))))))))))))))...................... (-19.56 = -19.90 + 0.33)

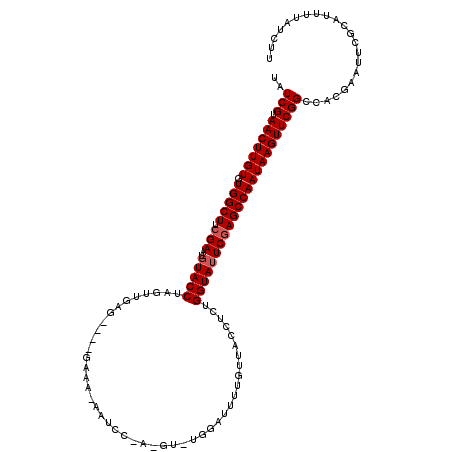

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:45 2006