| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,730,906 – 11,731,003 |
| Length | 97 |
| Max. P | 0.997663 |

| Location | 11,730,906 – 11,731,003 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -5.92 |
| Energy contribution | -6.96 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.29 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11730906 97 + 23771897 CAUCCUUUAUACACAUUUAUCUAUAUGUAUAUAUGUGUGUAGGCAAAUAUCUAUAUGCCUGAGUACACAUUUACUUGCGAAAGUAAGCGUUUGCAAG .......((((((.((......)).)))))).(((((((((((((.((....)).))))))..)))))))...((((((((.(....).)))))))) ( -24.10) >DroSec_CAF1 43776 95 + 1 CAGCCUUUAUACACAUUUAUCUAUAUGUGU--AUGUGUGUAGACAAAUAUCUAUAUGCCUGAGUACACAUUUACUUGCGAAAGUAAGCGUUUGCAAG (((.....((((((((........))))))--))(((((((((......)))))))))))).(((.((.(((((((....))))))).)).)))... ( -25.00) >DroSim_CAF1 38860 75 + 1 CAGCCUUUAUACACAUUUAUCUAUAUGUAU----------------------AUAUGCCUGAGUACACAUUUACUUGCGAAAGUAAGCGUUUGCAAG (((.(..((((((.((......)).)))))----------------------)...).))).(((.((.(((((((....))))))).)).)))... ( -14.70) >DroEre_CAF1 40127 87 + 1 CCUCCUUUAUACACAUUUAUUUGUAUAUGU----------AGGCAAAUAUCUAUAUGCCUGCGUACACAUUUACUUACGAAAGCCACCGUUCGCAAG .................((..(((.(((((----------(((((.((....)).)))))))))).)))..))(((.((((........)))).))) ( -19.10) >DroYak_CAF1 41001 83 + 1 ACCCCUUUAUACACAUAUAUUUAUAUAUGU--AUGUAUGAAGGCAAAUAUCU------------ACACAUUUACUUACGAAAGCAACCGUUUGCAAG ...((((((((((.((((((....))))))--.)))))))))).........------------..................((((....))))... ( -18.30) >consensus CAGCCUUUAUACACAUUUAUCUAUAUGUGU__AUGU_UG_AGGCAAAUAUCUAUAUGCCUGAGUACACAUUUACUUGCGAAAGUAAGCGUUUGCAAG ..........((((((........))))))...........................................((((((((.(....).)))))))) ( -5.92 = -6.96 + 1.04)


| Location | 11,730,906 – 11,731,003 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -8.24 |
| Energy contribution | -7.72 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
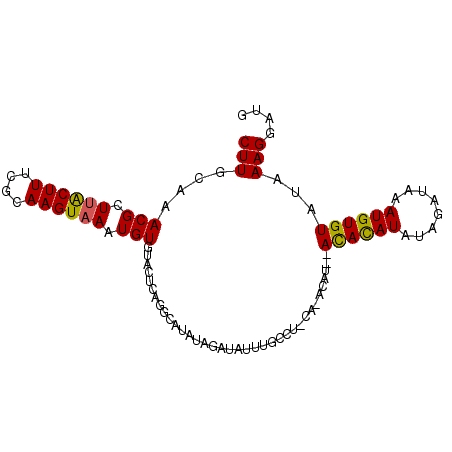
>3L_DroMel_CAF1 11730906 97 - 23771897 CUUGCAAACGCUUACUUUCGCAAGUAAAUGUGUACUCAGGCAUAUAGAUAUUUGCCUACACACAUAUAUACAUAUAGAUAAAUGUGUAUAAAGGAUG (((((..............)))))...((((((....(((((..........)))))..)))))).(((((((((......)))))))))....... ( -22.24) >DroSec_CAF1 43776 95 - 1 CUUGCAAACGCUUACUUUCGCAAGUAAAUGUGUACUCAGGCAUAUAGAUAUUUGUCUACACACAU--ACACAUAUAGAUAAAUGUGUAUAAAGGCUG .......((((((((((....))))))..)))).....(((.((((.(((((((((((.......--.......))))))))))).))))...))). ( -19.74) >DroSim_CAF1 38860 75 - 1 CUUGCAAACGCUUACUUUCGCAAGUAAAUGUGUACUCAGGCAUAU----------------------AUACAUAUAGAUAAAUGUGUAUAAAGGCUG .......((((((((((....))))))..)))).....(((...(----------------------((((((((......)))))))))...))). ( -16.20) >DroEre_CAF1 40127 87 - 1 CUUGCGAACGGUGGCUUUCGUAAGUAAAUGUGUACGCAGGCAUAUAGAUAUUUGCCU----------ACAUAUACAAAUAAAUGUGUAUAAAGGAGG ((((((.(((...((((....))))...)))...))))))..............(((----------..(((((((......)))))))..)))... ( -19.70) >DroYak_CAF1 41001 83 - 1 CUUGCAAACGGUUGCUUUCGUAAGUAAAUGUGU------------AGAUAUUUGCCUUCAUACAU--ACAUAUAUAAAUAUAUGUGUAUAAAGGGGU .(((((.....((((((....))))))...)))------------)).......((((.((((((--(.((((....)))).))))))).))))... ( -18.60) >consensus CUUGCAAACGCUUACUUUCGCAAGUAAAUGUGUACUCAGGCAUAUAGAUAUUUGCCU_CA_ACAU__ACACAUAUAGAUAAAUGUGUAUAAAGGAUG (((....(((.((((((....)))))).)))....................................((((((........))))))...))).... ( -8.24 = -7.72 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:38 2006