| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,728,816 – 11,729,008 |
| Length | 192 |
| Max. P | 0.816424 |

| Location | 11,728,816 – 11,728,917 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -17.44 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.81 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11728816 101 + 23771897 AUUCAAUUUGUCAAACCCAAUUCCCGCAAUUGACAGAAUGUGUAUUGCGCUACCCUUAUUUUUUGUAUU----UUUUUUUUUUGGUAGUAAUACAUAAAUUAAAU ......((((((((...............)))))))).((((((((..((((((...............----..........))))))))))))))........ ( -17.07) >DroSec_CAF1 41720 87 + 1 AUUCAAUUUGUCAAACCCAAUUCCCGCAAUUGACAGAAUGUGUAUUGCGCUACCCCU-----------------U-UUUUUUCUGUGGUAAUACAUAAAUUAAAU ......((((((((...............)))))))).((((((((((((.......-----------------.-........)).))))))))))........ ( -14.75) >DroSim_CAF1 36805 88 + 1 AUUCAAUUUGUCAAACCCAAUUCCCGCAAUUGACAGAAUGUGUAUUGCGCUACUCAU-----------------UUUUUUUUCUGUGGUAAUACAUAAAUUAAAU ......((((((((...............)))))))).((((((((..(((((....-----------------..........)))))))))))))........ ( -15.30) >DroEre_CAF1 38106 105 + 1 AUUCAAUUUGUCAAACCCAAUUCCCGCAAUGGACAGAAUGUGUAUUGCGCUACCAUUCCUGAUUUUAUUGCAAUUUUUUUUUUGCUGGUAAUACAUAAAUUUAAU ....(((((((.............(((((((.((.....)).))))))).(((((..............((((........)))))))))....))))))).... ( -17.23) >DroYak_CAF1 38870 95 + 1 AUUCAAUUUGUCAAAUGCAAUUCCCGCAAUUGACAGAAUGUGUAUUGCGCUAACAUACUUGAUUUUAU----------UUUUUGGUGGUAAUACAUAAAUUAAAU ......((((((((.(((.......))).)))))))).(((((((((((((((...............----------...))))).))))))))))........ ( -22.87) >consensus AUUCAAUUUGUCAAACCCAAUUCCCGCAAUUGACAGAAUGUGUAUUGCGCUACCCUU__U__UU_UAU______UUUUUUUUUGGUGGUAAUACAUAAAUUAAAU ......((((((((...............)))))))).((((((((((.......................................))))))))))........ (-12.61 = -12.81 + 0.20)

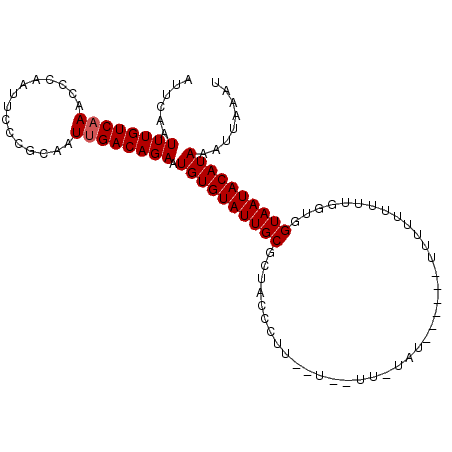
| Location | 11,728,917 – 11,729,008 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.38 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11728917 91 - 23771897 AUAAAUGCAAUACAUACAUAUAUGCAUACUAGUAUGUACAUAUGUGCCAGUCAGUGAGUGCGGACUCGUGUUCCA----------CCAAACCGAUUUCCAA ..............(((((((.((((((....)))))).)))))))..((((.((..(((.((((....))))))----------)...)).))))..... ( -20.70) >DroSec_CAF1 41807 99 - 1 AUAAAUGCAAUACAUAUAUGUACGACU-CU-ACAUGUACAUAUGUGCCAGUCAGUGAGUGCGGACCCGUGUUCCACUUGCUCCAACCAAAACGAUUUCCAA ............((((((((((((...-..-...))))))))))))......((..((((.((((....))))))))..)).................... ( -24.30) >DroSim_CAF1 36893 99 - 1 AUAAAUGCAAUACAUAUAUGUACGACU-AU-ACAUGUACAUAUGUGCCAGUCAGUGAGUGCGGACCCGUGUUCCACUUGCUCCAACCAAAACGAUUUCCAA ............((((((((((((...-..-...))))))))))))......((..((((.((((....))))))))..)).................... ( -24.30) >consensus AUAAAUGCAAUACAUAUAUGUACGACU_CU_ACAUGUACAUAUGUGCCAGUCAGUGAGUGCGGACCCGUGUUCCACUUGCUCCAACCAAAACGAUUUCCAA ............(((((((((((............))))))))))).......(.(((..((....))..))))........................... (-15.05 = -15.50 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:35 2006