| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,720,064 – 11,720,216 |
| Length | 152 |
| Max. P | 0.898652 |

| Location | 11,720,064 – 11,720,155 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -31.24 |
| Energy contribution | -32.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
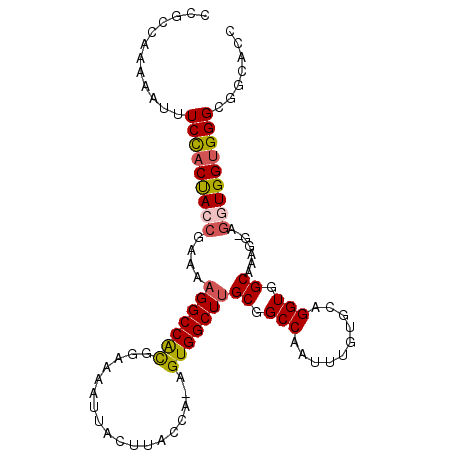
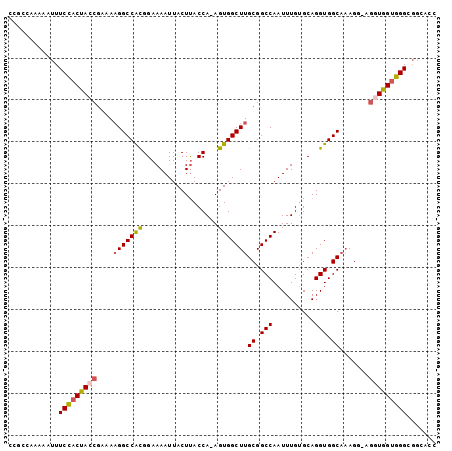
>3L_DroMel_CAF1 11720064 91 + 23771897 AGAGCUGAACUGGGCGUAAAGUGGGUGGUGGCAAGAGGGCGGGGCGUGGCGUGUUUCCGCCCACCACCUCCUUUGCCACCUGCACAAAUUG ...(((......))).....(((.(.(((((((.(((((.((((.(.((((......))))).)).))))))))))))))).)))...... ( -38.20) >DroSec_CAF1 32963 91 + 1 AGAGCUGAACUGGGCGUAUUGUGGGUGGUGGCCAGGGGGCGGGGCGUGGCGUGUUGCCGCCCACCACCUCCUUUGCCACCUGCACAAAUUG ...(((......)))...(((((.(.((((((.((((((.((((.(((((.....))))))).)).))))))..))))))).))))).... ( -45.20) >DroEre_CAF1 29244 87 + 1 AGAGCUGAACUGGGCGUAUAGUGGGUGGUGGAGAGCG---UGGGCGUGGCGUGGGGCCGCCC-CCACCUCCUUUGCCACCUGCACAAAUUG ...(((......))).....(..(((((..(((((.(---((((.(((((.....))))).)-)))))).)))..)))))..)........ ( -44.00) >DroYak_CAF1 29705 86 + 1 AGAGCUGAGCUGGGCGUAAAGUGGGUGGUGAAAAG-----GGGGCGUGGCAUGGUGCCGCCCACCACCUCCUUUGCCACCUGCACAAAUUG .....((.((.((..((((((..(((((((....(-----(.((((........)))).)))))))))..))))))..)).)).))..... ( -36.20) >consensus AGAGCUGAACUGGGCGUAAAGUGGGUGGUGGAAAG_G___GGGGCGUGGCGUGGUGCCGCCCACCACCUCCUUUGCCACCUGCACAAAUUG ...(((......))).....(..(((((..(..((.(((.((((.(((((.....))))))).)).))).)))..)))))..)........ (-31.24 = -32.80 + 1.56)


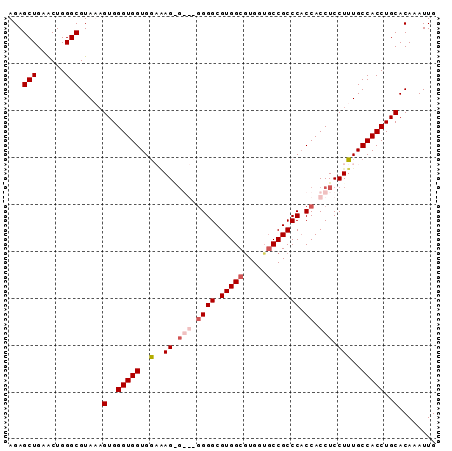
| Location | 11,720,064 – 11,720,155 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -26.14 |
| Energy contribution | -27.20 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11720064 91 - 23771897 CAAUUUGUGCAGGUGGCAAAGGAGGUGGUGGGCGGAAACACGCCACGCCCCGCCCUCUUGCCACCACCCACUUUACGCCCAGUUCAGCUCU ......(((..(((((((((((.((.((((((((......)))).))))))..))).))))))))...))).....((........))... ( -38.30) >DroSec_CAF1 32963 91 - 1 CAAUUUGUGCAGGUGGCAAAGGAGGUGGUGGGCGGCAACACGCCACGCCCCGCCCCCUGGCCACCACCCACAAUACGCCCAGUUCAGCUCU ....(((((..((((((..(((.(((((.(((.(((.....))).).))))))).))).))))))...)))))...((........))... ( -38.30) >DroEre_CAF1 29244 87 - 1 CAAUUUGUGCAGGUGGCAAAGGAGGUGG-GGGCGGCCCCACGCCACGCCCA---CGCUCUCCACCACCCACUAUACGCCCAGUUCAGCUCU ......(((..(((((....((((((((-(.(.(((.....))).).))))---).))))...)))))))).....((........))... ( -31.60) >DroYak_CAF1 29705 86 - 1 CAAUUUGUGCAGGUGGCAAAGGAGGUGGUGGGCGGCACCAUGCCACGCCCC-----CUUUUCACCACCCACUUUACGCCCAGCUCAGCUCU ....(((.((.((((..((((..(((((((((((((.....)))..)))).-----......))))))..)))).))))..)).))).... ( -33.01) >consensus CAAUUUGUGCAGGUGGCAAAGGAGGUGGUGGGCGGCAACACGCCACGCCCC___C_CUUGCCACCACCCACUAUACGCCCAGUUCAGCUCU ....(((.((.((((..((((..(((((((((((((.....)))..))))............))))))..)))).))))..)).))).... (-26.14 = -27.20 + 1.06)

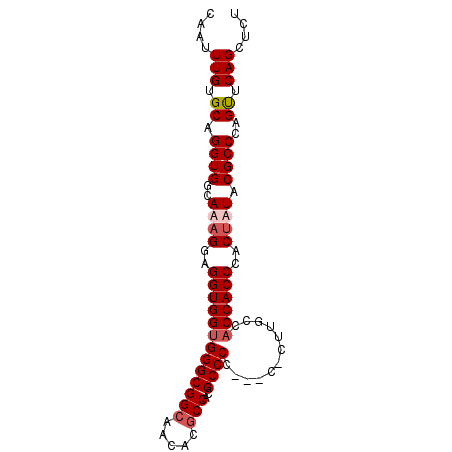
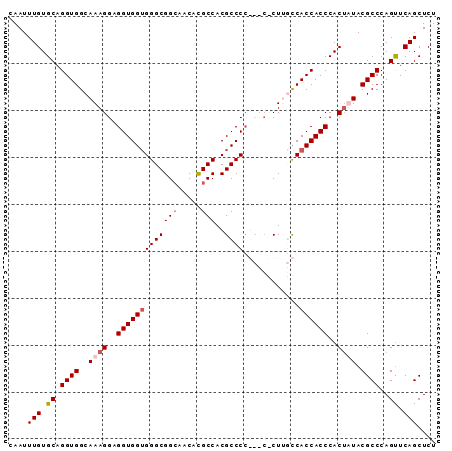
| Location | 11,720,116 – 11,720,216 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -27.06 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11720116 100 - 23771897 CCGCCAAAAAUUUCCACUACCGAAAAGGCCACGGAAAAUUACUUACCA-AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGG-AGGUGGUGGGCGGAAAC (((((........(((((.((....(((((((((.((.....)).)).-.)))))))((.(((..........))).))...))-.)))))..))))).... ( -35.30) >DroSec_CAF1 33015 100 - 1 CCGCCAAAAAUUUCCACUACCGAAAAGGCCACGGAAAAUUACUUACCA-AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGG-AGGUGGUGGGCGGCAAC (((((........(((((.((....(((((((((.((.....)).)).-.)))))))((.(((..........))).))...))-.)))))..))))).... ( -34.60) >DroSim_CAF1 27665 101 - 1 CCGCCAAAAAUUUCCACUACCGAAAAGGCCACGGAAAAUUACUUACCA-AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGGAAGGUGGUGGGCNNNNNN ((((((....(((((.....((...(((((((((.((.....)).)).-.))))))).))((((.(((....)))))))...))))).))))))........ ( -32.40) >DroEre_CAF1 29293 99 - 1 CCUCCAAAAAUUUCUACUAACGAAAAGGCCACGGAAAAUUACUUACCA-AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGG-AGGUGG-GGGCGGCCCC (((((...............((...(((((((((.((.....)).)).-.))))))).))((((.(((....)))))))...))-))).((-((....)))) ( -31.40) >DroYak_CAF1 29752 100 - 1 CCUCCAAAAAUUUCCACUACCGAAAAGGCCACGGAAAAUUACUUACCA-AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGG-AGGUGGUGGGCGGCACC ............(((((((((....(((((((((.((.....)).)).-.)))))))((.(((..........))).)).....-.)))))))))....... ( -31.50) >DroAna_CAF1 25677 95 - 1 ----UAAAAAUUUCCACCAACUGUACGGCCGUGGAAAAUUACUCACCAAAGUGGCUUGCGGCCAAUUUG-GCAGGUGGCUUAGA-A-GUGGUGGGUGGGCCG ----......(((((((...((....))..))))))).((((((((((((((.((((((.(......).-)))))).))))...-.-.)))))))))).... ( -32.80) >consensus CCGCCAAAAAUUUCCACUACCGAAAAGGCCACGGAAAAUUACUUACCA_AGUGGCUUGCGGCCAAUUUGUGCAGGUGGCAAAGG_AGGUGGUGGGCGGCACC ............(((((((((....(((((((..................)))))))((.(((..........))).)).......)))))))))....... (-27.06 = -27.34 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:32 2006