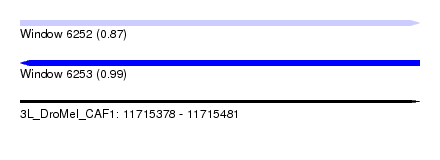
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,715,378 – 11,715,481 |
| Length | 103 |
| Max. P | 0.991448 |

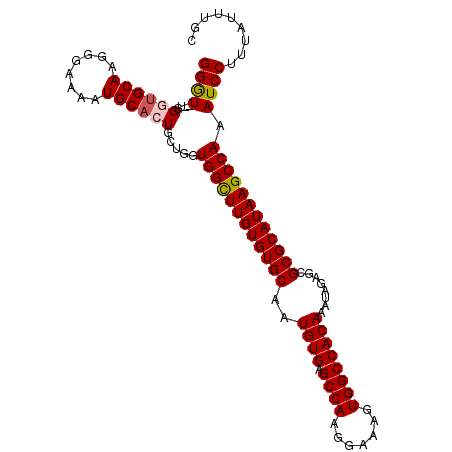
| Location | 11,715,378 – 11,715,481 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.53 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11715378 103 + 23771897 GCAAAUAAAGGAUUUGGCUUAUGCGCGCUCUAUUUGUGGCCACUUUCCUUGGCUCACAUUGCACACAAGCCAACAGCAGUGCAUUUUCCAUUGCACCCA--ACCC .........((..(((((((.((.(((.......((((((((.......)))).)))).))).)).)))))))..((((((.......))))))..)).--.... ( -29.80) >DroSim_CAF1 24412 103 + 1 GCAAAUAAAGGAUUUGGCUUAUGCGCGCUCUAUUUGUGGCCACUUUCCUUGGCUCACAUUGCACACAAGCCAACAGCAGUGCAUUUGCCAUUGCACCCA--AUCC .........(((((((((((.((.(((.......((((((((.......)))).)))).))).)).))))))...(((((((....).))))))....)--)))) ( -31.40) >DroEre_CAF1 24615 105 + 1 GCAAAUAAAGGAUUUGGCUUAUGCGCGCUCUAUUUGUGGCCACUUUCCUUGGCUCACAUUGCACACAAACCACCAGCACUGCAUUUUCCCUUGCACCCACCACCC ((((.....((.((((((....))(.((......((((((((.......)))).))))..)).).))))))....((...))........))))........... ( -20.90) >DroYak_CAF1 24647 103 + 1 GCAAAUAAAGGAUUUGGCUUAUGCGCGCUCUAUUUGUGGCCACUUUCCUUGGCUCACAUUGCACACAAACCACCAGCACUGCAUUUUCCCUUGC--CAGCCACCC .........((...((((..(((((.(((.....((((((((.......)))).))))(((....)))......))).).))))........))--)).)).... ( -21.70) >consensus GCAAAUAAAGGAUUUGGCUUAUGCGCGCUCUAUUUGUGGCCACUUUCCUUGGCUCACAUUGCACACAAACCAACAGCACUGCAUUUUCCAUUGCACCCA__ACCC .........((...((((((.((.(((.......((((((((.......)))).)))).))).)).))))))......(((((........)))))......)). (-21.46 = -22.53 + 1.06)

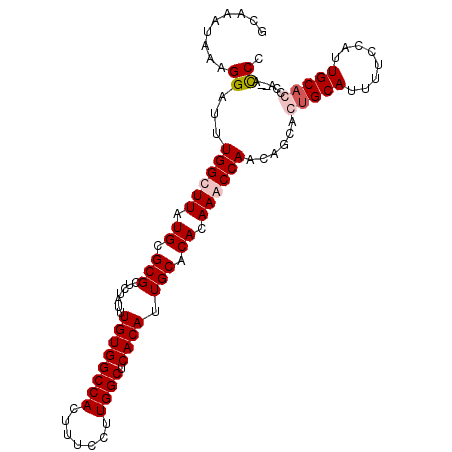
| Location | 11,715,378 – 11,715,481 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11715378 103 - 23771897 GGGU--UGGGUGCAAUGGAAAAUGCACUGCUGUUGGCUUGUGUGCAAUGUGAGCCAAGGAAAGUGGCCACAAAUAGAGCGCGCAUAAGCCAAAUCCUUUAUUUGC ((((--(.((((((.(....).)))))).....((((((((((((..((((.((((.......))))))))........)))))))))))))))))......... ( -39.30) >DroSim_CAF1 24412 103 - 1 GGAU--UGGGUGCAAUGGCAAAUGCACUGCUGUUGGCUUGUGUGCAAUGUGAGCCAAGGAAAGUGGCCACAAAUAGAGCGCGCAUAAGCCAAAUCCUUUAUUUGC ((((--(.((((((........)))))).....((((((((((((..((((.((((.......))))))))........)))))))))))))))))......... ( -39.00) >DroEre_CAF1 24615 105 - 1 GGGUGGUGGGUGCAAGGGAAAAUGCAGUGCUGGUGGUUUGUGUGCAAUGUGAGCCAAGGAAAGUGGCCACAAAUAGAGCGCGCAUAAGCCAAAUCCUUUAUUUGC ((((((.(((((((..(.......)..)))...((((((((((((..((((.((((.......))))))))........)))))))))))).)))).)))))).. ( -32.10) >DroYak_CAF1 24647 103 - 1 GGGUGGCUG--GCAAGGGAAAAUGCAGUGCUGGUGGUUUGUGUGCAAUGUGAGCCAAGGAAAGUGGCCACAAAUAGAGCGCGCAUAAGCCAAAUCCUUUAUUUGC ((((.((..--(((..(.......)..)))..))(((((((((((..((((.((((.......))))))))........)))))))))))..))))......... ( -35.50) >consensus GGGU__UGGGUGCAAGGGAAAAUGCACUGCUGGUGGCUUGUGUGCAAUGUGAGCCAAGGAAAGUGGCCACAAAUAGAGCGCGCAUAAGCCAAAUCCUUUAUUUGC ((((....((((((........)))))).....((((((((((((..((((.((((.......))))))))........)))))))))))).))))......... (-33.06 = -33.38 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:25 2006