
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,712,823 – 11,712,944 |
| Length | 121 |
| Max. P | 0.910039 |

| Location | 11,712,823 – 11,712,923 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.49 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11712823 100 + 23771897 UCUCCGCCUCCUUUGAGGUCCUUGCCACACACCACCACCCCCUCCCCCCCCUGCGCUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCA .....(((((....)))))....((((.........................(((.((...)).)))..((((((((.......))))).)))))))... ( -21.60) >DroSec_CAF1 26045 79 + 1 UCGCCGCCUCCUUUGAGGUCCUUGCCACAACC---------------------CGGUGCCCCACCGCUUGGUACGUGUAUGCGUCACGUCGCCUGGUUCA .....(((((....)))))....((((.....---------------------(((((...)))))...((((((((.......))))).)))))))... ( -22.30) >DroSim_CAF1 21975 79 + 1 UCGCCGCCUCCUUUGAGGUCCUUGCCACACCC---------------------CGGUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCA .....(((((....)))))....((((.....---------------------(((((...)))))...((((((((.......))))).)))))))... ( -24.30) >consensus UCGCCGCCUCCUUUGAGGUCCUUGCCACACCC_____________________CGGUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCA .....(((((....)))))....((((..........................(((((...)))))...((((((((.......))))).)))))))... (-21.45 = -21.57 + 0.11)
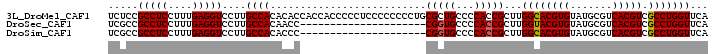

| Location | 11,712,846 – 11,712,944 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
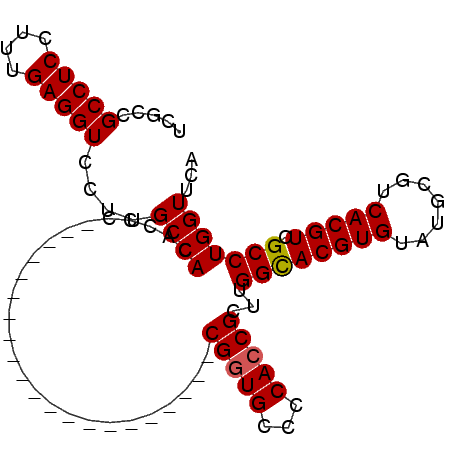
>3L_DroMel_CAF1 11712846 98 + 23771897 GCCACACACCACCACCCCCUCCCCCCCCUGCGCUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCAUCACCAGGACCUCCUGCCCAC ..........((((...............(((.((...)).)))..((((((((.......))))).))))))).......((((....))))..... ( -20.70) >DroSec_CAF1 26068 77 + 1 GCCACAACC---------------------CGGUGCCCCACCGCUUGGUACGUGUAUGCGUCACGUCGCCUGGUUCAUCACCAGGACCUCCUGCCCAA ((((.....---------------------(((((...)))))...((((((((.......))))).))))))).......((((....))))..... ( -21.40) >DroSim_CAF1 21998 77 + 1 GCCACACCC---------------------CGGUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCAUCACCAGGACCUCCUGCCCAA ((((.....---------------------(((((...)))))..))))(((((.......)))))..((((((.....))))))............. ( -23.60) >consensus GCCACACCC_____________________CGGUGCCCCACCGCUUGGCACGUGUAUGCGUCACGUCGCCUGGUUCAUCACCAGGACCUCCUGCCCAA ((((..........................(((((...)))))...((((((((.......))))).))))))).......((((....))))..... (-20.55 = -20.67 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:23 2006