
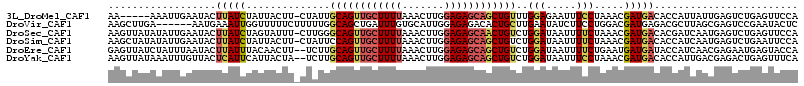
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,704,260 – 11,704,403 |
| Length | 143 |
| Max. P | 0.998577 |

| Location | 11,704,260 – 11,704,363 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.91 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
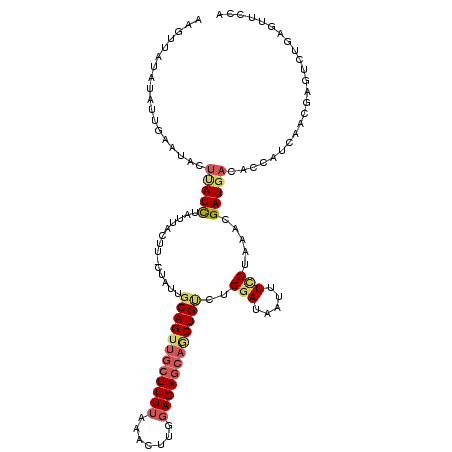
>3L_DroMel_CAF1 11704260 103 + 23771897 UGUGAUGCUGAUGUAAGCU----AUAGGAGUAUUCUAAAA-----AAAUUGAAUACUUAUCUAUUACUU-CUAUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUUUGGAGA .((((((((......))).----....((((((((.....-----.....))))))))....)))))((-(((..((((((((((((.......)))))))))))).))))). ( -31.80) >DroVir_CAF1 21493 106 + 1 CACAAGCCCAACGUGAGUUUGUCAUUAAUUUAAUC-ACAAGCUUGA------AAUGAAAUUGGUUUUUCUUUUUGGCAGCUGAUUUGUGCAUUGGAGAGACACUGCUUGAAUA ..(((((.((...(((((((((.((((...)))).-))))))))).------..)).....(((.(((((((...(((.(......))))...))))))).)))))))).... ( -25.10) >DroSec_CAF1 17541 108 + 1 UGUGAUGCUGAGGUAAGCC----AUAAAAGUAUUCUAAAAGUUAUAUAUUGAAUACUUAUCUAGUAUUU-CUUGGGCAGUUGCUUUUAAACUUGGAGAGCAACUGUCUGGAUA .(.(((((((.((....))----....((((((((...............))))))))...))))))).-)(..(((((((((((((.......)))))))))))))..)... ( -33.86) >DroSim_CAF1 13012 108 + 1 UGUGAUGCUGAGGUAAGCU----AUUAAAGUAUUCUAAAAGCUAUAUAUUGAAUACUUAUCUAUUACUU-CUAUUCCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUA .((((((((......))).----....((((((((...............))))))))....))))).(-(((...(((((((((((.......)))))))))))..)))).. ( -27.36) >DroEre_CAF1 13527 107 + 1 UGUGAUGCUGAGGUAAACU----UUAACAGUAUAAUUAGAGUUAUCUAUUUAAUACUUAUUUACAACUU--UCUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUA (((((((.(((((....))----))).))((((((.((((....)))).)).))))....)))))...(--((..((((((((((((.......))))))))))))..))).. ( -27.50) >DroYak_CAF1 13604 107 + 1 UGUGAUGCUGAUGUAAGCU----UUAACAGAGUUCUGAAAGUUAUAAAUUUGUUACUCAUUCAUUACUA--UCUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUA .((((((.(((.(((((((----((..(((....))))))))..........)))))))..))))))((--(((.((((((((((((.......))))))))))))..))))) ( -30.00) >consensus UGUGAUGCUGAGGUAAGCU____AUAAAAGUAUUCUAAAAGUUAUAUAUUGAAUACUUAUCUAUUACUU_CUAUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUA ............................(((((((...............)))))))..................((((((((((((.......))))))))))))....... (-12.54 = -13.91 + 1.37)



| Location | 11,704,294 – 11,704,403 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.75 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -13.41 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11704294 109 + 23771897 AA-----AAAUUGAAUACUUAUCUAUUACUU-CUAUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUUUGGAGAAUUUCCUAAACGAUGACACCAUUAUUGAGUCUGAGUUCCA ..-----.........(((((.((....(((-(....((((((((((((.......))))))))))))..))))...........((((((.....))))))))..))))).... ( -23.70) >DroVir_CAF1 21530 109 + 1 AAGCUUGA------AAUGAAAUUGGUUUUUCUUUUUGGCAGCUGAUUUGUGCAUUGGAGAGACACUGCUUGAAUAUCUUCCUGGACGAUGAGACGCUUAGCGAGUCCGAAUACUC ..(((.((------((.((((......)))))))).))).(((((..(((.(((((..(((......))).....(((....))))))))..))).)))))((((......)))) ( -19.50) >DroSec_CAF1 17575 114 + 1 AAGUUAUAUAUUGAAUACUUAUCUAGUAUUU-CUUGGGCAGUUGCUUUUAAACUUGGAGAGCAACUGUCUGGAUAAUUUUCUAAACGAUGACACGAUCAAUGAGUCUGAGUUCCA (((((((.....(((((((.....)))))))-.(..(((((((((((((.......)))))))))))))..))))))))....(((...(((.((.....)).)))...)))... ( -29.00) >DroSim_CAF1 13046 114 + 1 AAGCUAUAUAUUGAAUACUUAUCUAUUACUU-CUAUUCCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUAAUUUUCUAAACGAUGACACCAUCAAUGAGUCUGAAUUCCA ............((((((((((........(-(((...(((((((((((.......)))))))))))..)))).............((((....)))).))))))....)))).. ( -22.70) >DroEre_CAF1 13561 113 + 1 GAGUUAUCUAUUUAAUACUUAUUUACAACUU--UCUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUAAUUUUCUGAAUGAUGAUACCAUCAACGAGAAUGAGUACCA ...............(((((((((.(....(--((..((((((((((((.......))))))))))))..)))............(((((....)))))..).)))))))))... ( -29.50) >DroYak_CAF1 13638 113 + 1 AAGUUAUAAAUUUGUUACUCAUUCAUUACUA--UCUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUAAUUUCCUAAACGAUGACACCAUUGACGAGACUGAGUUUCA ............((..(((((.((.....((--(((.((((((((((((.......))))))))))))..)))))..........(((((....)))))....)).)))))..)) ( -31.50) >consensus AAGUUAUAUAUUGAAUACUUAUCUAUUACUU_CUAUUGCAGUUGCUUUUAAACUUGGAGAGCAGCUGUCUGGAUAAUUUCCUAAACGAUGACACCAUCAACGAGUCUGAGUUCCA ..................(((((..............((((((((((((.......))))))))))))..(((.....))).....)))))........................ (-13.41 = -14.17 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:20 2006