
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,670,412 – 11,670,510 |
| Length | 98 |
| Max. P | 0.650648 |

| Location | 11,670,412 – 11,670,510 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
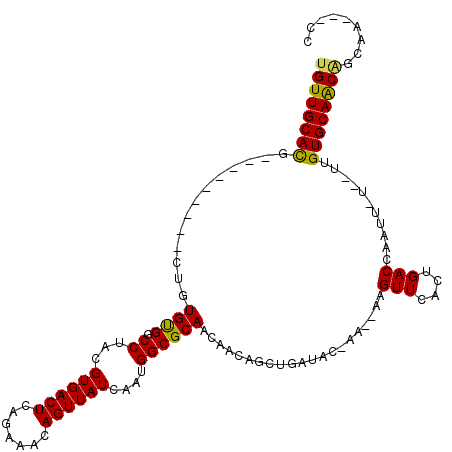
>3L_DroMel_CAF1 11670412 98 - 23771897 UGUUGCAUG---------GUGUGCGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACAC------AUACGGA--AAGUUCAGUGACCAAAA-U--UUGUGCAGCAGCCA-AACG ((((((((.---------(((((((.((...((((((.......))))))....))))).))))------.(((.((--....)).))).......-.--..))))))))....-.... ( -25.50) >DroVir_CAF1 2042 108 - 1 UGUUGCACGUUGCG----CUGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACAACAGCUGCUUC-AGCUGAGUUCACUGACCAAUU-U--UCGUGCAACAGCAA---CC ((((((((((((((----(((((((...)))((((((.......))))))..))))))))))..((((((...)-)))))................-.--..))))))))....---.. ( -36.70) >DroPse_CAF1 4949 102 - 1 UGUUGCACA---------CUGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACGUGCGAGUAUAC-GA--AAGUUCAGUGACCAAUUUU--CUGUGCAGCAGC---AGCA (((((((((---------..((((((((...((((((.......))))))....)))((.....))...)))))-((--(((((........))))))--))))))))))..---.... ( -28.70) >DroGri_CAF1 3439 110 - 1 UGUUGCACGUUGUUGUGGCCGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACAACAGCUGCUUC-AA--AAGUUCACUGACCAAUU-U--UAGUGCAACGGCAA---CC .(((((.(((((..(..((.((.(((((...((((((.......))))))....))).))...)).))..)...-..--.....((((((......-)--))))))))))))))---). ( -33.40) >DroMoj_CAF1 4526 108 - 1 UGUUGCACGUUGCU----CUGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACAACAGCUGCUUC-AA--AAGUUCACUGACCAAUU-CCGUAGUGCAACUGCAA---CC .(((((((((((((----(((.((.((....)).)).))))....((((....)))))))))..(((..(((..-..--.)))...))).......-.....))))))).....---.. ( -30.00) >DroPer_CAF1 4957 105 - 1 UGUUGCACA---------CUGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACGUGCGAGUAUAC-GA--AAGUUCAGUGACCAAUUUU--CUGUGCAGCAGCAGCAGCA (((((((((---------..((((((((...((((((.......))))))....)))((.....))...)))))-((--(((((........))))))--))))))))))((....)). ( -29.90) >consensus UGUUGCACG_________CUGUGUGGCCUACGUGACUCAGAAACAGUUAUCAAUGGCGCAACAACAGCUGAUAC_AA__AAGUUCACUGACCAAUU_U__UUGUGCAACAGCAA___CC ((((((((.............((((.((...((((((.......))))))....)))))).....................(((....)))...........))))))))......... (-20.15 = -19.68 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:09 2006