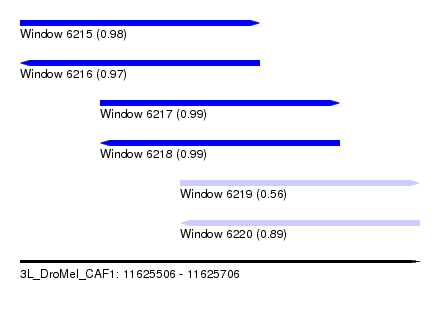
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,625,506 – 11,625,706 |
| Length | 200 |
| Max. P | 0.993949 |

| Location | 11,625,506 – 11,625,626 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -34.35 |
| Energy contribution | -34.48 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11625506 120 + 23771897 UGCCACGCGUAGCAUGCAGUCAUCUUUAGCUUGGAUUCCCUUGUUCACUCUGCUCCUUGGCCACCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAACAG ..(((.((..((.(((....)))))...)).))).......((((((....(((((((((((((.(((.((((.((((....)))).)))).))))))).....))))))))))))))). ( -43.40) >DroSec_CAF1 438 112 + 1 --------GUAGAAUGCAGUCAUCUUUAGUUUGGAUUCCCUUGUUCACUCUGCUCCUUGGCCAGCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCUAAGGAGCUGAGCAG --------...((((.(((...........))).))))...((((((....((((((((((((.((((.((((.((((....)))).)))).))))))).....))))))))))))))). ( -38.10) >DroSim_CAF1 561 120 + 1 UGCCACGCGUAGAAUGCAGUCAUCUUUAGCUUGGAUUUCCUUGUUCACUCUUCUCCUUGGCCACCAGGUUUCAGCCAAGCUUCUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAG ..(((.((..(((.((....)))))...)).))).......((((((.....((((((((((((.(((.((((.(((......))).)))).))))))).....)))))))).)))))). ( -39.40) >DroYak_CAF1 547 120 + 1 GGCCACGCGCAGAAUGCAGUCAUCUUUAGCUUGGAUUCCCUUGUUCACCCUGCUCCUUGGCCACCAGGUUUCAUCUCAAUUUUUGGAUGAGUCCUGUGGAAAAUUCCAGGAGCAGAGCAG (((...(((.....))).))).......((((((((......)))))..((((((((.((((((.(((.(((((((........))))))).)))))))......))))))))))))).. ( -38.40) >consensus UGCCACGCGUAGAAUGCAGUCAUCUUUAGCUUGGAUUCCCUUGUUCACUCUGCUCCUUGGCCACCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAG ...........((((.(((.(.......).))).))))...((((((....((((((((((((.((((.((((.(((......))).)))).))))))).....))))))))))))))). (-34.35 = -34.48 + 0.13)


| Location | 11,625,506 – 11,625,626 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -31.24 |
| Energy contribution | -32.92 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11625506 120 - 23771897 CUGUUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGGUGGCCAAGGAGCAGAGUGAACAAGGGAAUCCAAGCUAAAGAUGACUGCAUGCUACGCGUGGCA .(((((((((((((((.....(((((((.((((.((((....)))).)))).))).)))))))))))))....)))))).((..(((.........)))..))...(((((....))))) ( -43.20) >DroSec_CAF1 438 112 - 1 CUGCUCAGCUCCUUAGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGCUGGCCAAGGAGCAGAGUGAACAAGGGAAUCCAAACUAAAGAUGACUGCAUUCUAC-------- ..((((.(((((((.(.....(((((((.((((.((((....)))).)))).)))).)))).))))))).))))(((...((..(((.........)))..))...)))...-------- ( -34.40) >DroSim_CAF1 561 120 - 1 CUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAGAAGCUUGGCUGAAACCUGGUGGCCAAGGAGAAGAGUGAACAAGGAAAUCCAAGCUAAAGAUGACUGCAUUCUACGCGUGGCA ..((((..((((((((.....(((((((.((((.((((....)))).)))).))).))))))))))))..))))............(((.((....((((....))))....)).))).. ( -40.00) >DroYak_CAF1 547 120 - 1 CUGCUCUGCUCCUGGAAUUUUCCACAGGACUCAUCCAAAAAUUGAGAUGAAACCUGGUGGCCAAGGAGCAGGGUGAACAAGGGAAUCCAAGCUAAAGAUGACUGCAUUCUGCGCGUGGCC ..(((((((((((((......(((((((..(((((((.....)).)))))..))).)))))).)))))))))))............(((.((...(((((....)).)))..)).))).. ( -40.70) >consensus CUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGGUGGCCAAGGAGCAGAGUGAACAAGGGAAUCCAAGCUAAAGAUGACUGCAUUCUACGCGUGGCA .......(((((((((.....(((((((.((((.((((....)))).)))).)))).))))))))))))((((((.....((..(((.........)))..)).)))))).......... (-31.24 = -32.92 + 1.69)

| Location | 11,625,546 – 11,625,666 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -48.50 |
| Consensus MFE | -42.84 |
| Energy contribution | -42.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11625546 120 + 23771897 UUGUUCACUCUGCUCCUUGGCCACCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAACAGGGCAGGACUACAGGCAGCUGCUUGGAUGACAGCCAUCAGU (((((((....(((((((((((((.(((.((((.((((....)))).)))).))))))).....))))))))))))))))(((.(.....(((((....))))).....).)))...... ( -48.60) >DroSec_CAF1 470 120 + 1 UUGUUCACUCUGCUCCUUGGCCAGCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCUAAGGAGCUGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCUGUCAGU ((((((.(((.((((((((((((.((((.((((.((((....)))).)))).))))))).....))))))))).)))..)))))).......(((((((((......).))))))))... ( -51.80) >DroSim_CAF1 601 120 + 1 UUGUUCACUCUUCUCCUUGGCCACCAGGUUUCAGCCAAGCUUCUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAGGGCAGGAUUACAGGCCGCUGCUUGGAUGACAGCUGUCAGU ............((((((((((((.(((.((((.(((......))).)))).))))))).....))))))))(..((((((((..(....)..))).)))))..).((((....)))).. ( -48.20) >DroYak_CAF1 587 120 + 1 UUGUUCACCCUGCUCCUUGGCCACCAGGUUUCAUCUCAAUUUUUGGAUGAGUCCUGUGGAAAAUUCCAGGAGCAGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCCGUCAGU ..((((.(((((((((((((((((.(((.(((((((........))))))).)))))))......)))))....))))))))..))))..(((((....)))))..((((....)))).. ( -45.40) >consensus UUGUUCACUCUGCUCCUUGGCCACCAGGUUUCAGCCAAGCUUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCCGUCAGU (((((((....((((((((((((.((((.((((.(((......))).)))).))))))).....))))))))))))))))(((.(.....(((((....))))).....).)))...... (-42.84 = -42.65 + -0.19)

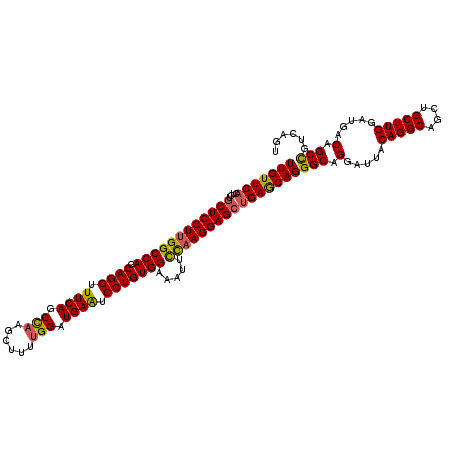

| Location | 11,625,546 – 11,625,666 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -37.47 |
| Energy contribution | -38.60 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11625546 120 - 23771897 ACUGAUGGCUGUCAUCCAAGCAGCUGCCUGUAGUCCUGCCCUGUUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGGUGGCCAAGGAGCAGAGUGAACAA ((.(..((((((.......))))))..).))..........(((((((((((((((.....(((((((.((((.((((....)))).)))).))).)))))))))))))....)))))). ( -45.50) >DroSec_CAF1 470 120 - 1 ACUGACAGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCAGCUCCUUAGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGCUGGCCAAGGAGCAGAGUGAACAA ((.(.(((((((.......))))))).).))...........((((.(((((((.(.....(((((((.((((.((((....)))).)))).)))).)))).))))))).))))...... ( -41.30) >DroSim_CAF1 601 120 - 1 ACUGACAGCUGUCAUCCAAGCAGCGGCCUGUAAUCCUGCCCUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAGAAGCUUGGCUGAAACCUGGUGGCCAAGGAGAAGAGUGAACAA .......(((((.......)))))(((..(....)..)))..((((..((((((((.....(((((((.((((.((((....)))).)))).))).))))))))))))..))))...... ( -43.20) >DroYak_CAF1 587 120 - 1 ACUGACGGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCUGCUCCUGGAAUUUUCCACAGGACUCAUCCAAAAAUUGAGAUGAAACCUGGUGGCCAAGGAGCAGGGUGAACAA ((.(.(((((((.......))))))).).))......((((((((((((..(((((....))))((((..(((((((.....)).)))))..)))))..))...))))))))))...... ( -45.30) >consensus ACUGACAGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAAGCUUGGCUGAAACCUGGUGGCCAAGGAGCAGAGUGAACAA ((.(.(((((((.......))))))).).))...........((((.(((((((((.....(((((((.((((.((((....)))).)))).)))).)))))))))))).))))...... (-37.47 = -38.60 + 1.13)



| Location | 11,625,586 – 11,625,706 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -32.04 |
| Energy contribution | -31.35 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
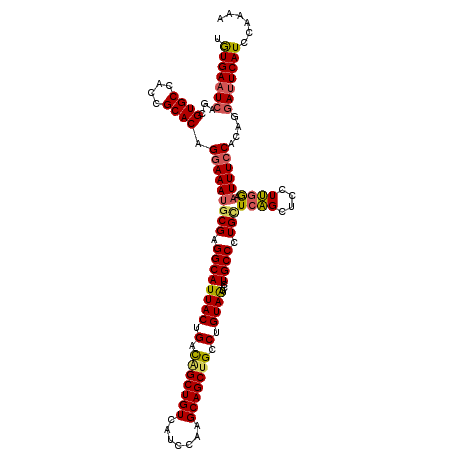
>3L_DroMel_CAF1 11625586 120 + 23771897 UUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAACAGGGCAGGACUACAGGCAGCUGCUUGGAUGACAGCCAUCAGUAAUGCCUCGCAUUUCCUGUGCGGUGGCACGCUGAUUCACA ....(((....))).(((((...((((((.(((((....((......)).....))))).)))))).........(((((..((((((((((.....)))))).)))).)))))))))). ( -43.60) >DroSec_CAF1 510 120 + 1 UUUUGGAUGAAUCCUGUGGAAAUUCUAAGGAGCUGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCUGUCAGUAAUGCCACGCAUUUCCUGUGCGGUGGCACGCUGAUUCACG .......((((((((((...(.(((....))).)..))))(((...(((((.(((((((((......).)))))))).)))))(((((((((.....)))).)))))..))))))))).. ( -43.60) >DroSim_CAF1 641 120 + 1 UUCUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAGGGCAGGAUUACAGGCCGCUGCUUGGAUGACAGCUGUCAGUAAUGCCACGCAUUUCCUGUGCGGUGGCACGCUGAUUCAUA ..(((......((((.(((.....))))))).(..((((((((..(....)..))).)))))..)....)))..((((((..((((((((((.....)))).)))))).))))))..... ( -46.50) >DroYak_CAF1 627 120 + 1 UUUUGGAUGAGUCCUGUGGAAAAUUCCAGGAGCAGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCCGUCAGUAAUGCCUCGGAUUUUCUGUGCGGUGGCACUCUUAUUCACA ...(((((((((((((.(((...))))))))((...(((.((.((((((..(((((..((((..((((.....)))))))).)))))..)))))))).)))....))...)))))))).. ( -38.90) >consensus UUUUGGAUGAAUCCUGUGGAAAUUCCAAGGAGCUGAGCAGGGCAGGAUUACAGGCAGCUGCUUGGAUGACAGCCGUCAGUAAUGCCACGCAUUUCCUGUGCGGUGGCACGCUGAUUCACA ....(((....)))((((((.......(((((....((..((((..(((.(((((....)))))((((.....)))))))..))))..))..)))))((((....)))).....)))))) (-32.04 = -31.35 + -0.69)


| Location | 11,625,586 – 11,625,706 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -30.65 |
| Energy contribution | -31.03 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11625586 120 - 23771897 UGUGAAUCAGCGUGCCACCGCACAGGAAAUGCGAGGCAUUACUGAUGGCUGUCAUCCAAGCAGCUGCCUGUAGUCCUGCCCUGUUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAA .(((((((...(((((..((((.......)))).)))))..(((.(((..((..((((((.(((((...(((....)))......))))).))))))))..)))))))))))))...... ( -38.10) >DroSec_CAF1 510 120 - 1 CGUGAAUCAGCGUGCCACCGCACAGGAAAUGCGUGGCAUUACUGACAGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCAGCUCCUUAGAAUUUCCACAGGAUUCAUCCAAAA .(((((((...((((....)))).(((((((((.((((((((.(.(((((((.......))))))).).))))...)))).))).....((....))))))))....)))))))...... ( -38.60) >DroSim_CAF1 641 120 - 1 UAUGAAUCAGCGUGCCACCGCACAGGAAAUGCGUGGCAUUACUGACAGCUGUCAUCCAAGCAGCGGCCUGUAAUCCUGCCCUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAGAA .(((((((...(((((((.(((.......))))))))))..(((..(((((.......(((((.(((..(....)..)))))))))))))...(((.....)))))))))))))...... ( -39.61) >DroYak_CAF1 627 120 - 1 UGUGAAUAAGAGUGCCACCGCACAGAAAAUCCGAGGCAUUACUGACGGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCUGCUCCUGGAAUUUUCCACAGGACUCAUCCAAAA ((((.......((((....)))).((((((((((((.(((((.(.(((((((.......))))))).).))))))))((........))...))).))))))))))(((....))).... ( -33.80) >consensus UGUGAAUCAGCGUGCCACCGCACAGGAAAUGCGAGGCAUUACUGACAGCUGUCAUCCAAGCAGCUGCCUGUAAUCCUGCCCUGCUCAGCUCCUUGGAAUUUCCACAGGAUUCAUCCAAAA .(((((((...((((....)))).(((((((((.((((((((.(.(((((((.......))))))).).))))...)))).)))((((....)))).))))))....)))))))...... (-30.65 = -31.03 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:53 2006