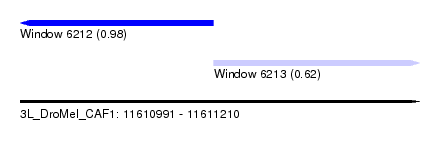
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,610,991 – 11,611,210 |
| Length | 219 |
| Max. P | 0.979375 |

| Location | 11,610,991 – 11,611,097 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
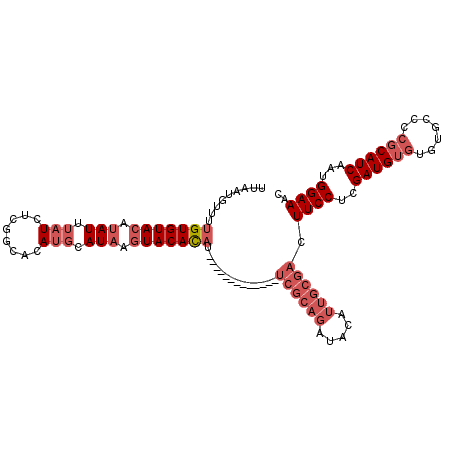
>3L_DroMel_CAF1 11610991 106 - 23771897 GCUCACAUACCGACUUCUGGCUAUAUAUAUAUUUACUCAAACAGUGGGUUAUGAACCCGCCAUACCCCAAAGAAAACAACAACUUGUUUGCUUAACACACUGCAGA ((...............((((.............(((.....)))((((.....)))))))).......(((.((((((....)))))).)))........))... ( -16.00) >DroSec_CAF1 104628 102 - 1 GCUCACAUACCGACUUCUGGGUAU----AUAUUCACACAAACAGUGGGUUAUGAAGCCACGAUACCCCAAAUGAAACAACAACUUGUGGGCUUAACCCACUGUAGA ......(((((........)))))----............(((((((((((...((((((((.....................)))).)))))))))))))))... ( -27.40) >DroSim_CAF1 105070 99 - 1 GCUCACAUACCGACUUCUGGGUAU----AUAUUUACACAAACAGUGGGUUAUGAAGCCGCGAUAUCCCAAAUGAAAC---AAUUUGUGGGCUUAACCCACUGCAGA ......(((((........)))))----.............((((((((((...((((.((......(((((.....---.))))))))))))))))))))).... ( -26.40) >DroEre_CAF1 95422 86 - 1 GCUCACAUACUGACUUCUGGGUAC----AUAUUUACUCAAACAGUGGGUUAUAAG----------------GGAAACAACAACGUGUUUGCUUAACCCACUGCACA .................((((((.----.....))))))..((((((((((...(----------------(.(((((......))))).)))))))))))).... ( -24.70) >DroYak_CAF1 109957 86 - 1 GCUCACAUACUGACUUCUGGGUGA----AUAUUUGCUCAAACAGUGGGUUAUAAA----------------GAAAACAACAACGUGUUUGAUUAACCCACUGCAGA .........(((.....((((..(----....)..))))..((((((((((....----------------..(((((......)))))...))))))))))))). ( -25.10) >consensus GCUCACAUACCGACUUCUGGGUAU____AUAUUUACUCAAACAGUGGGUUAUGAA_CC_C_AUA_CCCAAAGGAAACAACAACUUGUUUGCUUAACCCACUGCAGA .......((((........))))..................((((((((((.........................................)))))))))).... (-13.26 = -13.26 + -0.00)

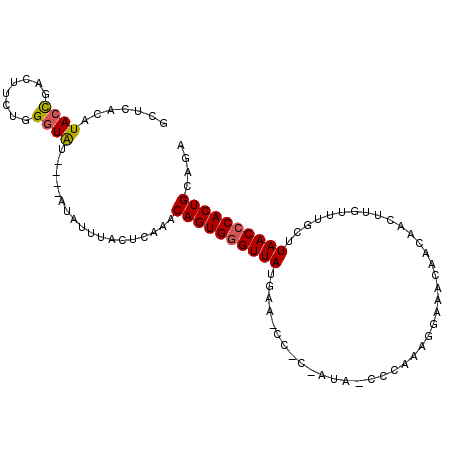
| Location | 11,611,097 – 11,611,210 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -17.34 |
| Energy contribution | -19.37 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
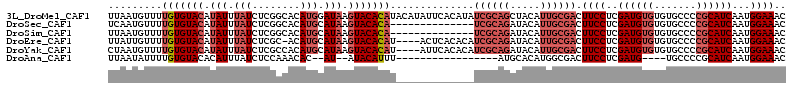
>3L_DroMel_CAF1 11611097 113 + 23771897 UUAAUGUUUUGUGUACAUAUUUAUCUCGGCACAUGGAUAAGUACACAUACAUAUUCACAUAUCGCAGCUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC ...((((..(((((((.(((((((........))))))).))))))).)))).........((((((.....)))))).((((..((((((.......))))))...)))).. ( -30.70) >DroSec_CAF1 104730 99 + 1 UCAAUGUUUUGUGUACAUAUUUAUCUCGGCACAUGCAUAAGUACACA--------------UCGCAGAUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC .........(((((((.(((.(((........))).))).)))))))--------------((((((.....)))))).((((..((((((.......))))))...)))).. ( -24.00) >DroSim_CAF1 105169 99 + 1 UUAAUGUUUUGUGUACAUAUUUAUCUCGGCACAUGCAUAAGUACACA--------------UCGCAGAUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC .........(((((((.(((.(((........))).))).)))))))--------------((((((.....)))))).((((..((((((.......))))))...)))).. ( -24.00) >DroEre_CAF1 95508 108 + 1 UUAUUGUUUUGUGUACAUAUUUAUCUCGC-ACAUGCAUAAGUACACAU----ACUCACACAUCGCAGAUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC ((((((...(((((((.(((.(((.....-..))).))).))))))).----.(.((((((((((((.....))))((.....)))))))))).)........)))))).... ( -24.40) >DroYak_CAF1 110043 109 + 1 CUAAUGUUUUGUGUACAUAUUUAUCUCGCCACAUGCAUAAGUACACAU----AUUCACACAUCGCAGAUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC ...((((.......))))..........(((.((((....((((((((----(((......((((((.....)))))).......)))))))))))...))))...))).... ( -24.62) >DroAna_CAF1 114868 88 + 1 UUAAUAUUUUGUGUACACAUUUAUCUCCAAACAC--AU--AUACAUUU-----------------AUGCACAUGGCGACUUCCUCGAUG----UGCCCCGCAUCAAUGGAAAC .........(((....)))....((.(((....(--((--(......)-----------------)))....))).)).((((..((((----((...))))))...)))).. ( -14.70) >consensus UUAAUGUUUUGUGUACAUAUUUAUCUCGGCACAUGCAUAAGUACACAU_____________UCGCAGAUACAUUGCGACUUCCUCGAUGUGUGUGCCCCGCAUCAAUGGAAAC .........(((((((.(((.(((........))).))).)))))))..............((((((.....)))))).((((..((((((.......))))))...)))).. (-17.34 = -19.37 + 2.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:46 2006