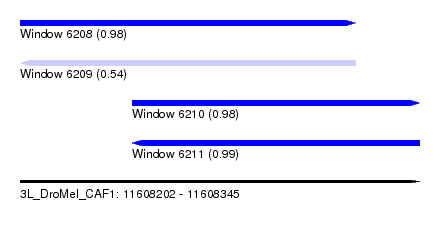
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,608,202 – 11,608,345 |
| Length | 143 |
| Max. P | 0.993879 |

| Location | 11,608,202 – 11,608,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.07 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -20.63 |
| Energy contribution | -20.51 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11608202 120 + 23771897 UCUUGACUUUUACGUUUGGCGUAUGGAUUCUGUGGGUCGUCGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAAAUCACCGCAAUUAGCAUACAAUUCAAGUUCAAGUUCAAACU .............((((((........(((....((((..(.....)..))))....)))..(((((((((((((..........((.....))......))))))))))))))))))). ( -27.89) >DroSec_CAF1 101819 105 + 1 -------AUUUACGAUUGGC--------UCUGCGGGUCGUCGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACU -------((((.((((((((--------...(((((((...)))))))..))))))))))))(((((((((((((.........(((.....))).....)))))))))))))....... ( -35.34) >DroSim_CAF1 102305 112 + 1 UCUUGACUUUUACGAUUGGC--------UCUGCGGGUCGUCGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUGCAAUUCAAGUUCAAGUUUAAACU ............((((((((--------...(((((((...)))))))..))))))))....(((((((((((((......((.(((.....)))))...)))))))))))))....... ( -36.30) >DroEre_CAF1 92628 118 + 1 GCUUGACUUUUACGAUUGCUAUAUGG-CUCUGUAGGUCAUCGAUCUGGUGGGUAAUUGAAAUACCUGAUCUUGAAAAAGAUCGCUGGUAUUAGCAUACAAU-CAAGUUCAAGUUUAAGCU (((((((((..(((((((...(((((-(((..((((((...))))))..))))......(((((((((((((....)))))))..))))))..))))))))-)..))..))).)))))). ( -34.50) >DroYak_CAF1 107065 119 + 1 UCCUGACUUUUACGAUUGGUAUAUGG-CUCUGUGGGUCGUCGAUCUGGUGGGUAAUUGAAAAACUUGAACUUGAAAAAGAUCGCUUGUAUUAGCAUACAAUUCAAGUUCAAGUUAAAACU ..((.(((.....(((((....((((-((.....))))))))))).))).)).........((((((((((((((....((.(((......)))))....))))))))))))))...... ( -31.10) >consensus UCUUGACUUUUACGAUUGGC_UAUGG__UCUGUGGGUCGUCGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACU ............((((((((............((((((...))))))...))))))))....(((((((((((((.........................)))))))))))))....... (-20.63 = -20.51 + -0.12)


| Location | 11,608,202 – 11,608,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.07 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11608202 120 - 23771897 AGUUUGAACUUGAACUUGAAUUGUAUGCUAAUUGCGGUGAUUUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCGACGACCCACAGAAUCCAUACGCCAAACGUAAAAGUCAAGA ..((((((((((((((((((..((.(((.....)))...))....))))))))))))).......(((((.....((((...........).))).....))))).........))))). ( -27.30) >DroSec_CAF1 101819 105 - 1 AGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCGACGACCCGCAGA--------GCCAAUCGUAAAU------- .......(((((((((((((..(..(((.....))).....)...)))))))))))))(((((.((((((.......((..((...))..))--------)))))).).))))------- ( -24.71) >DroSim_CAF1 102305 112 - 1 AGUUUAAACUUGAACUUGAAUUGCAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCGACGACCCGCAGA--------GCCAAUCGUAAAAGUCAAGA .......((((((((((((((..(.(((.....))))..)).....))))))))))))......((((((.......((..((...))..))--------)))))).............. ( -26.11) >DroEre_CAF1 92628 118 - 1 AGCUUAAACUUGAACUUG-AUUGUAUGCUAAUACCAGCGAUCUUUUUCAAGAUCAGGUAUUUCAAUUACCCACCAGAUCGAUGACCUACAGAG-CCAUAUAGCAAUCGUAAAAGUCAAGC .((((..((((.....((-(((((..(((......)))((((((....))))))((((...((.(((........))).))..))))......-.......)))))))...)))).)))) ( -23.30) >DroYak_CAF1 107065 119 - 1 AGUUUUAACUUGAACUUGAAUUGUAUGCUAAUACAAGCGAUCUUUUUCAAGUUCAAGUUUUUCAAUUACCCACCAGAUCGACGACCCACAGAG-CCAUAUACCAAUCGUAAAAGUCAGGA .(....((((((((((((((..(..((((......))))..)...))))))))))))))...).........((.(((..((((.........-...........))))....))).)). ( -24.45) >consensus AGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCGACGACCCACAGA__CCAUA_GCCAAUCGUAAAAGUCAAGA .......(((((((((((((.....((((......))))......))))))))))))).((((.((((((.......((...........))........)))))).).)))........ (-17.04 = -17.84 + 0.80)

| Location | 11,608,242 – 11,608,345 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -14.11 |
| Energy contribution | -15.51 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11608242 103 + 23771897 CGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAAAUCACCGCAAUUAGCAUACAAUUCAAGUUCAAGUUCAAACUCUAUUAGAAGAGCUAACUGGUAA----------------- ...((((((((....((((...(((((((((((((..........((.....))......)))))))))))))))))...))))))))...............----------------- ( -23.69) >DroSec_CAF1 101844 115 + 1 CGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACUCUAUUUGAAGAGCUAACUGGUACUGCAACAAUAU-----A .....(((.(((((.(((....(((((((((((((.........(((.....))).....))))))))))))).....((((......)))).))).)))))..))).......-----. ( -25.84) >DroSim_CAF1 102337 115 + 1 CGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUGCAAUUCAAGUUCAAGUUUAAACUUUACCAGAAGAGCUAACUGGUACUGCAACAAUAU-----A .....((((.((..........(((((((((((((......((.(((.....)))))...)))))))))))))........((((((.........))))))..))))))....-----. ( -26.60) >DroEre_CAF1 92667 119 + 1 CGAUCUGGUGGGUAAUUGAAAUACCUGAUCUUGAAAAAGAUCGCUGGUAUUAGCAUACAAU-CAAGUUCAAGUUUAAGCUUUAUCAGAAGAGCUAAUUGGUACUGCAUGAAUAUUCUAUA .......(((((..(((.((((((((((((((....)))))))..)))))).((((((...-..(((((((((....))))........))))).....))).))).).)))..))))). ( -24.90) >DroYak_CAF1 107104 104 + 1 CGAUCUGGUGGGUAAUUGAAAAACUUGAACUUGAAAAAGAUCGCUUGUAUUAGCAUACAAUUCAAGUUCAAGUUAAAACUUUCUUAGAAUAUU-AUAUCAUAAAG--------------- .(((.(((((.......((((((((((((((((((....((.(((......)))))....)))))))))))))).....))))......))))-).)))......--------------- ( -24.52) >consensus CGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACUUUAUUAGAAGAGCUAACUGGUACUGCA__AAUAU_____A ...((((((((....((((...(((((((((((((.........................)))))))))))))))))...))))))))................................ (-14.11 = -15.51 + 1.40)



| Location | 11,608,242 – 11,608,345 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11608242 103 - 23771897 -----------------UUACCAGUUAGCUCUUCUAAUAGAGUUUGAACUUGAACUUGAAUUGUAUGCUAAUUGCGGUGAUUUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCG -----------------...((((((((((((......))))))...(((((((((((((..((.(((.....)))...))....))))))))))))).....))))))........... ( -29.40) >DroSec_CAF1 101844 115 - 1 U-----AUAUUGUUGCAGUACCAGUUAGCUCUUCAAAUAGAGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCG .-----...(((((......((((((((((((......))))))...(((((((((((((..(..(((.....))).....)...))))))))))))).....))))))..))))).... ( -31.00) >DroSim_CAF1 102337 115 - 1 U-----AUAUUGUUGCAGUACCAGUUAGCUCUUCUGGUAAAGUUUAAACUUGAACUUGAAUUGCAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCG .-----...(((((((..((((((.........))))))........((((((((((((((..(.(((.....))))..)).....))))))))))))..........)).))))).... ( -32.80) >DroEre_CAF1 92667 119 - 1 UAUAGAAUAUUCAUGCAGUACCAAUUAGCUCUUCUGAUAAAGCUUAAACUUGAACUUG-AUUGUAUGCUAAUACCAGCGAUCUUUUUCAAGAUCAGGUAUUUCAAUUACCCACCAGAUCG .............((.((((((....((((..........))))....((((((...(-(((((............))))))...))))))....)))))).))................ ( -19.30) >DroYak_CAF1 107104 104 - 1 ---------------CUUUAUGAUAU-AAUAUUCUAAGAAAGUUUUAACUUGAACUUGAAUUGUAUGCUAAUACAAGCGAUCUUUUUCAAGUUCAAGUUUUUCAAUUACCCACCAGAUCG ---------------...........-..........((((.....((((((((((((((..(..((((......))))..)...))))))))))))))))))................. ( -21.50) >consensus U_____AUAUU__UGCAGUACCAGUUAGCUCUUCUAAUAAAGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCG ...............................................(((((((((((((.....((((......))))......)))))))))))))...................... (-16.88 = -16.72 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:44 2006