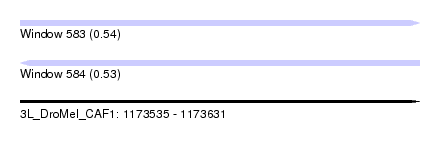
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,173,535 – 1,173,631 |
| Length | 96 |
| Max. P | 0.536561 |

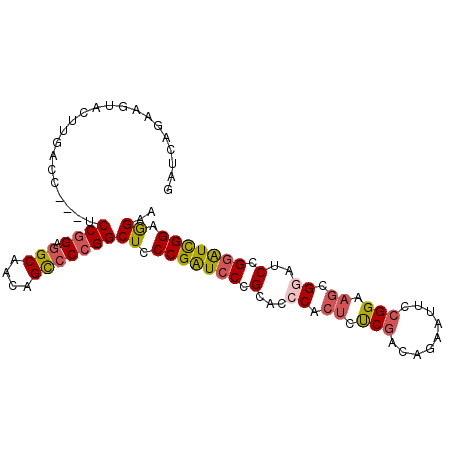
| Location | 1,173,535 – 1,173,631 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
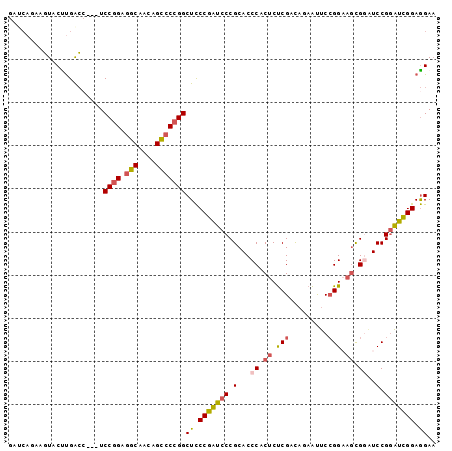
>3L_DroMel_CAF1 1173535 96 + 23771897 GAUCACAGGUACUUGACC---UCCGGAGGCAAUAGUCCCGGCUCCCGAUCCCGCACCCACUCUCGACAGAAUUCCGGAAGUGGAUCCGGAUCGGAGGAA ......((((.....)))---)((((.(((....)))))))(..(((((((.(...(((((..((.........))..)))))..).)))))))..).. ( -34.80) >DroSec_CAF1 57379 96 + 1 GAUCAGAAGUACUUGACC---UCCGGAGGCAAUAGCCCCGGCUCCCGAUCCCGCACCCACUCUCGACAGAAUUCCGGAAGCGGGUCCGGAUCGGAGGAA ..((((......))))((---(((((.(((....)))))))...(((((((.(.((((.((..((.........))..)).))))).)))))))))).. ( -38.90) >DroSim_CAF1 55877 96 + 1 GAUCAGAAGUACUUGACC---UCCGGAGGCAAUAGCCCCGGCUCCCGAUCCCGCACCCACUCUCGACAGAAUUCCGGAAGCGGGUCCGGGUCGGAGGAA ..((((......))))((---(((((.(((....)))))))...((((.((((.((((.((..((.........))..)).)))).))))))))))).. ( -40.20) >DroEre_CAF1 55687 88 + 1 --------GUGCCUGACA---UCCGGGGGCAACAGCCCCGGCUCCCGAUCCCGCACCCACUCUCGCCAAAACUCCGGAAGCGCAUCCGCAUCGGAGGAA --------((((..((..---..(((((((..........))))))).))..))))...............((((((..(((....))).))))))... ( -27.50) >DroYak_CAF1 59758 96 + 1 GAGCACAAGUACCUGACC---UCCGGAGGCAACAGCCCCGGCUCCCGAUCCCGCACCCACUCUCGCCAAAACUCCGGCAGCGCAUCCGGAUCGGAGGAA ...........(((....---.((((.(((....)))))))...((((((((((..........(((........))).))).....)))))))))).. ( -29.70) >DroAna_CAF1 62225 99 + 1 GACCAGCGCUCCUCGGGCGGAUCCAGAAGCAACAGCCCCGGCUUCCGGUCCCGCACCCACUCCCGCUCCAGUUCGGGCUUCGGUUCCGGAUUGGAAGAC .....(((......((((((..((.(((((..........))))).))..)))).))......)))((((((((((((....).))))))))))).... ( -40.90) >consensus GAUCAGAAGUACUUGACC___UCCGGAGGCAACAGCCCCGGCUCCCGAUCCCGCACCCACUCUCGACAGAAUUCCGGAAGCGGAUCCGGAUCGGAGGAA ......................((((.(((....)))))))((.(((((((.(...((.((.(((.........))).)).))..).))))))).)).. (-22.06 = -22.57 + 0.50)

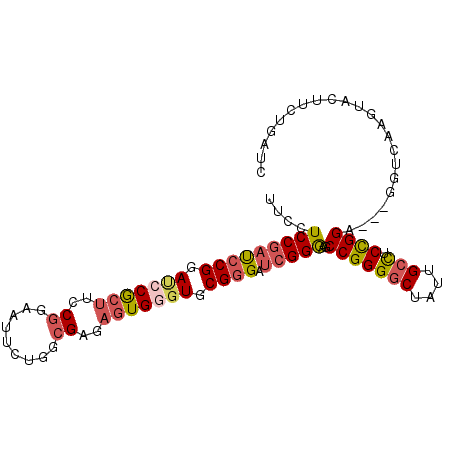
| Location | 1,173,535 – 1,173,631 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -31.52 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1173535 96 - 23771897 UUCCUCCGAUCCGGAUCCACUUCCGGAAUUCUGUCGAGAGUGGGUGCGGGAUCGGGAGCCGGGACUAUUGCCUCCGGA---GGUCAAGUACCUGUGAUC ..(..(((((((.((((((((..(((.......)))..))))))).).)))))))..).(((((((...((((....)---)))..))).))))..... ( -39.20) >DroSec_CAF1 57379 96 - 1 UUCCUCCGAUCCGGACCCGCUUCCGGAAUUCUGUCGAGAGUGGGUGCGGGAUCGGGAGCCGGGGCUAUUGCCUCCGGA---GGUCAAGUACUUCUGAUC ..(..(((((((.((((((((..(((.......)))..))))))).).)))))))..)(((((((....))).)))).---(((((........))))) ( -46.40) >DroSim_CAF1 55877 96 - 1 UUCCUCCGACCCGGACCCGCUUCCGGAAUUCUGUCGAGAGUGGGUGCGGGAUCGGGAGCCGGGGCUAUUGCCUCCGGA---GGUCAAGUACUUCUGAUC ..(..((((((((.(((((((..(((.......)))..))))))).)))).))))..)(((((((....))).)))).---(((((........))))) ( -48.00) >DroEre_CAF1 55687 88 - 1 UUCCUCCGAUGCGGAUGCGCUUCCGGAGUUUUGGCGAGAGUGGGUGCGGGAUCGGGAGCCGGGGCUGUUGCCCCCGGA---UGUCAGGCAC-------- ....((((((.(.(((.((((..((.........))..)))).)).).).)))))).((((((((....)))))..(.---...).)))..-------- ( -33.80) >DroYak_CAF1 59758 96 - 1 UUCCUCCGAUCCGGAUGCGCUGCCGGAGUUUUGGCGAGAGUGGGUGCGGGAUCGGGAGCCGGGGCUGUUGCCUCCGGA---GGUCAGGUACUUGUGCUC ..((((((((((.(((.(((((((((....)))))...)))).)).).)))))))...(((((((....))).)))))---))...((((....)))). ( -40.10) >DroAna_CAF1 62225 99 - 1 GUCUUCCAAUCCGGAACCGAAGCCCGAACUGGAGCGGGAGUGGGUGCGGGACCGGAAGCCGGGGCUGUUGCUUCUGGAUCCGCCCGAGGAGCGCUGGUC ((.((((....(((.(((.(..((((........))))..).)))(((((.((((((((..........)))))))).)))))))).)))).))..... ( -39.90) >consensus UUCCUCCGAUCCGGAUCCGCUUCCGGAAUUCUGGCGAGAGUGGGUGCGGGAUCGGGAGCCGGGGCUAUUGCCUCCGGA___GGUCAAGUACUUCUGAUC ....(((((((((.(((((((..((.........))..))))))).)))).)))))..(((((((....))).))))...................... (-31.52 = -32.33 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:54 2006