| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,579,043 – 11,579,140 |
| Length | 97 |
| Max. P | 0.883145 |

| Location | 11,579,043 – 11,579,140 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
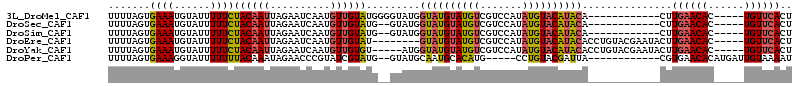
>3L_DroMel_CAF1 11579043 97 + 23771897 UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUAUGGGGUAUGGUAUGUAUGUCGUCCAUAUGUACAUACA------------CUUGAACAC-----UGUUCACU ....((((((.(((....(((((....)))))((((.((.((((((.((((((.(((.....))))))))).).))))))------------)))))))).-----..)))))) ( -24.30) >DroSec_CAF1 74613 95 + 1 UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUAUG--GUAUGGUAUGUAUGUCGUCCAUAUGUACAUACA------------CUUGAACAC-----UGUUCACU ....((((((.(((....(((((....)))))((((.((.(((((--((((((.(((.....)))))))))...))))))------------)))))))).-----..)))))) ( -22.50) >DroSim_CAF1 76343 95 + 1 UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUAUG--GUAUGGUAUGUAUGUCGUCCAUAUGUACAUACA------------CUUGAACAC-----UGUUCACU ....((((((.(((....(((((....)))))((((.((.(((((--((((((.(((.....)))))))))...))))))------------)))))))).-----..)))))) ( -22.50) >DroEre_CAF1 65882 101 + 1 UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUAU--------GUAUGUAUGUCGUCCAUAUGUACAUACACCUGUACGAAUACUUGAACAC-----UGUUCACU ...(((((....(((((((((((....))))).......(((((--------(((((((((.....)))))))))))))).......)))))).....)))-----))...... ( -24.10) >DroYak_CAF1 80747 104 + 1 UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUGU-----AUGGUAUGUAUGUCGUCCAUAUGUACAUACACCUGUACGAAUACUUGAACAC-----UGUUCACU ....((((((.(((....(((((....)))))((((.((..(((-----((((..((((((((((....))).))))))).)))))))...))))))))).-----..)))))) ( -24.40) >DroPer_CAF1 64024 95 + 1 UUUUAGUGAAAGGUAUUUUUUUACAAAUAGAACCCGUAUCGUAUG--GUAUGCAAUGCACAUG-----CCUGUACGAUUA------------CGUGAACACAUGAUUGUAAAAU .....(((....(((......)))........(.(((((((((((--(((((.......))))-----)).)))))).))------------)).)..)))............. ( -20.90) >consensus UUUUAGUGAAAUGUAUUUUUCUACAAUUAGAAUCAAUGUUGUAUG__GUAUGGUAUGUAUGUCGUCCAUAUGUACAUACA____________CUUGAACAC_____UGUUCACU .......((((......))))((((((..........)))))).........((((((((((.......))))))))))...............((((((......)))))).. (-12.25 = -12.62 + 0.36)
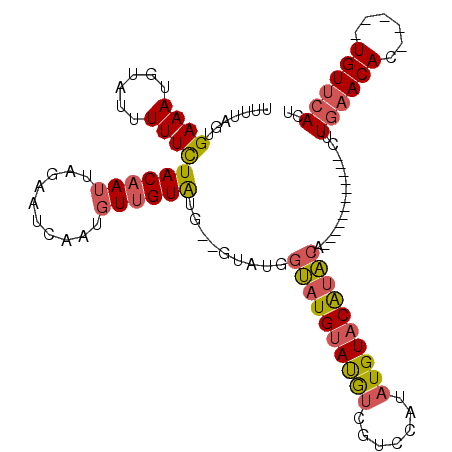
| Location | 11,579,043 – 11,579,140 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -8.35 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11579043 97 - 23771897 AGUGAACA-----GUGUUCAAG------------UGUAUGUACAUAUGGACGACAUACAUACCAUACCCCAUACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ((((((..-----((((((((.------------((((((....(((((............)))))...))))))...))))(((((....)))))..))))..)))))).... ( -20.90) >DroSec_CAF1 74613 95 - 1 AGUGAACA-----GUGUUCAAG------------UGUAUGUACAUAUGGACGACAUACAUACCAUAC--CAUACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ((((((((-----(((((....------------.(((((((..((((.....))))..)).)))))--.....))))))).(((((....)))))........)))))).... ( -20.60) >DroSim_CAF1 76343 95 - 1 AGUGAACA-----GUGUUCAAG------------UGUAUGUACAUAUGGACGACAUACAUACCAUAC--CAUACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ((((((((-----(((((....------------.(((((((..((((.....))))..)).)))))--.....))))))).(((((....)))))........)))))).... ( -20.60) >DroEre_CAF1 65882 101 - 1 AGUGAACA-----GUGUUCAAGUAUUCGUACAGGUGUAUGUACAUAUGGACGACAUACAUAC--------AUACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ((((((((-----(((((...(((....)))....(((((((..((((.....))))..)))--------))))))))))).(((((....)))))........)))))).... ( -25.00) >DroYak_CAF1 80747 104 - 1 AGUGAACA-----GUGUUCAAGUAUUCGUACAGGUGUAUGUACAUAUGGACGACAUACAUACCAU-----ACACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ((((((((-----(((((...(((....)))..(((((((((..((((.....))))..)).)))-----))))))))))).(((((....)))))........)))))).... ( -25.50) >DroPer_CAF1 64024 95 - 1 AUUUUACAAUCAUGUGUUCACG------------UAAUCGUACAGG-----CAUGUGCAUUGCAUAC--CAUACGAUACGGGUUCUAUUUGUAAAAAAAUACCUUUCACUAAAA .((((((((....(...((.((------------((.(((((..((-----.((((.....)))).)--).)))))))))))..)...)))))))).................. ( -18.80) >consensus AGUGAACA_____GUGUUCAAG____________UGUAUGUACAUAUGGACGACAUACAUACCAUAC__CAUACAACAUUGAUUCUAAUUGUAGAAAAAUACAUUUCACUAAAA ..((((((......))))))..............(((((((...........))))))).......................(((((....))))).................. ( -8.35 = -8.93 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:33 2006