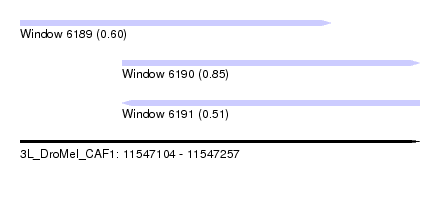
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,547,104 – 11,547,257 |
| Length | 153 |
| Max. P | 0.854718 |

| Location | 11,547,104 – 11,547,223 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -29.21 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11547104 119 + 23771897 GUUUUGUAGUCCCUGGACCGUGUUUCAUCCAUAUCCUGUUCAAUAGGCGCCUUUUGCGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGACGCCUUUUUCGC ....(((.(((....)))...........................(((((((((((.(((((.....))))).)))))))))))))).....((((........))))........... ( -29.70) >DroSec_CAF1 45653 119 + 1 GUUUUGUAGUCCCUGGACGGUGUUUCAUCCAUAUCCUGUUGAAUAGGCACCUUUUGCGGACUAUUAAUGCCUUUGAAAGGUGCUGCAUUCAUGUCCUGUCGAAUGGGCGCCUUUUUCGC ((...(((((((.(..(.(((((((.((.((........)).))))))))).)..).)))))))....))....((((((((((.(((((..........))))))))))))))).... ( -35.90) >DroSim_CAF1 44070 119 + 1 GUUUUGUAGUCCCUGGACUGUGUUUCAUCCAUAUCCUGUUGAAUGGGCACCUUUUGGGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGGCGUCUUUUUCGC ......(((((....)))))....................((((((((((((((..((((((.....))))))..))))))))).)))))((((((........))))))......... ( -41.70) >consensus GUUUUGUAGUCCCUGGACCGUGUUUCAUCCAUAUCCUGUUGAAUAGGCACCUUUUGCGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGGCGCCUUUUUCGC ........(((....))).(((.................(((((.((((((((((..((((.......))))..))))))))))..)))))(((((........))))).......))) (-29.21 = -29.43 + 0.23)

| Location | 11,547,143 – 11,547,257 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -30.73 |
| Energy contribution | -30.40 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11547143 114 + 23771897 UCAAUAGGCGCCUUUUGCGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGACGCCUUUUUCGCAGUUUUGAAUUGCGGUCCCUCUGGGGGUUUUUUC ......(((((((((((.(((((.....))))).)))))))))))........((((........)))).......(((((((.....)))))))((((....))))....... ( -37.10) >DroSec_CAF1 45692 114 + 1 UGAAUAGGCACCUUUUGCGGACUAUUAAUGCCUUUGAAAGGUGCUGCAUUCAUGUCCUGUCGAAUGGGCGCCUUUUUCGCAGUUUUGAAGUGCUGAGCCUCUGGUGGUUUUUUC ......((((((((((..((.(.......)))...))))))))))(((((((....(((.((((.((....))..)))))))...)).))))).(((((......))))).... ( -31.80) >DroSim_CAF1 44109 114 + 1 UGAAUGGGCACCUUUUGGGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGGCGUCUUUUUCGCAGUUUUGAAGUGCGGACCCUCUGGUGGUUUUUUC .((((((((((((((..((((((.....))))))..))))))))).)))))((((((........))))))......((((.(.....).))))(((((....).))))..... ( -43.30) >consensus UGAAUAGGCACCUUUUGCGGACUAUUAAAGUCUUUGAAAGGUGCUGCAUUCAUGUCCUGUUUAAUGGGCGCCUUUUUCGCAGUUUUGAAGUGCGGACCCUCUGGUGGUUUUUUC ......((((((((((..((((.......))))..))))))))))(((((((.((((........))))((.......)).....)))).)))....((......))....... (-30.73 = -30.40 + -0.33)



| Location | 11,547,143 – 11,547,257 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11547143 114 - 23771897 GAAAAAACCCCCAGAGGGACCGCAAUUCAAAACUGCGAAAAAGGCGUCCAUUAAACAGGACAUGAAUGCAGCACCUUUCAAAGACUUUAAUAGUCCGCAAAAGGCGCCUAUUGA .......(((.....)))...(((.((((.....((.......))((((........)))).))))))).((.(((((....((((.....))))....))))).))....... ( -27.30) >DroSec_CAF1 45692 114 - 1 GAAAAAACCACCAGAGGCUCAGCACUUCAAAACUGCGAAAAAGGCGCCCAUUCGACAGGACAUGAAUGCAGCACCUUUCAAAGGCAUUAAUAGUCCGCAAAAGGUGCCUAUUCA (((..................(((.((((...(((((((...(....)..)))).)))....))))))).((((((((....(((.......)))....))))))))...))). ( -25.10) >DroSim_CAF1 44109 114 - 1 GAAAAAACCACCAGAGGGUCCGCACUUCAAAACUGCGAAAAAGACGCCCAUUAAACAGGACAUGAAUGCAGCACCUUUCAAAGACUUUAAUAGUCCCCAAAAGGUGCCCAUUCA .......((......))((((.............(((.......)))..........)))).((((((..((((((((....((((.....))))....)))))))).)))))) ( -28.00) >consensus GAAAAAACCACCAGAGGGUCCGCACUUCAAAACUGCGAAAAAGGCGCCCAUUAAACAGGACAUGAAUGCAGCACCUUUCAAAGACUUUAAUAGUCCGCAAAAGGUGCCUAUUCA .......((......))....(((.((((.....(((.......)))((........))...))))))).((((((((....((((.....))))....))))))))....... (-20.71 = -20.93 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:25 2006