
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,511,590 – 11,511,681 |
| Length | 91 |
| Max. P | 0.842775 |

| Location | 11,511,590 – 11,511,681 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -21.79 |
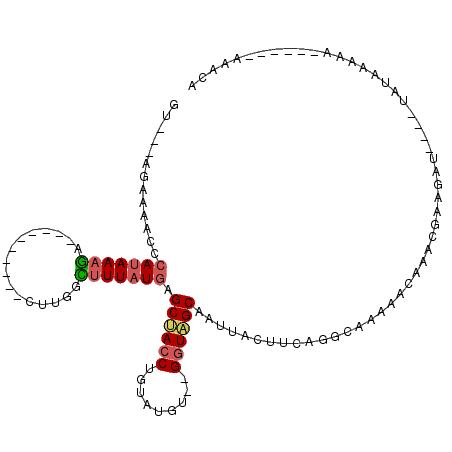
| Consensus MFE | -8.90 |
| Energy contribution | -9.12 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
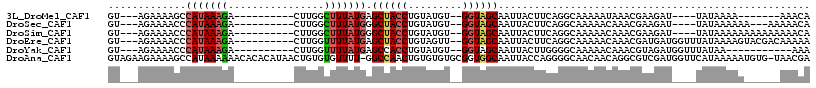
>3L_DroMel_CAF1 11511590 91 + 23771897 GU---AGAAAAGCCAUAAAGA----------CUUGGCUUUAUGAGCUACCUGUAUGU--GGUAGCAAUUACUUCAGGCAAAAAUAAACGAAGAU----UAUAAAA-------AAACA ..---...(((((((......----------..)))))))....((((((.......--)))))).....((((..............))))..----.......-------..... ( -20.64) >DroSec_CAF1 17586 95 + 1 GU---AGAAAACCCAUAAAGA----------CUUGGCUUUAUGGGCUACCUGUAUGU--GGUAGCAAUUACUUCAGGCAAAAACAAACGAAGAU----UAUAAAAAA---AAAAACA ..---.......((((((((.----------.....))))))))((((((.......--)))))).....((((..............))))..----.........---....... ( -20.94) >DroSim_CAF1 15948 98 + 1 GU---AGAAAACCCAUAAAGA----------CUUGGCUUUAUGGGCUACCUGUAUGU--GGUAGCAAUUACUUCAGGCAAAAACAAACGAAGAU----UAUAAAAAAAAAAAAAACA ..---.......((((((((.----------.....))))))))((((((.......--)))))).....((((..............))))..----................... ( -20.94) >DroEre_CAF1 11967 102 + 1 GU---AGAAAACCCAUAAAGA----------CUUGGUUUUAUGAGCUACCUGUAGUU--GGUAGCAAUUACUUCAGGCAAAAACAAACGAUGAUGGUUUAUAAAAGUACGACAAAAA ((---(......(((......----------..)))(((((((((((((((((((((--(....)))))))...)))......((.....)).)))))))))))).)))........ ( -18.00) >DroYak_CAF1 16250 91 + 1 GU---AGAAAACCCAUAAAGA----------CUUGGUUUUAUGAGCCACCUGUAUGU--GGUAGCAAUUACUUGGGGCAAAAACAAACGUAGAUGGUUUAUAA-----------AAA ..---.......(((......----------..)))((((((((((((.((((.(((--.((..(((....)))..))....))).))..)).))))))))))-----------)). ( -18.40) >DroAna_CAF1 29159 115 + 1 GUAGAAGAAAAGCCAUAAAAAACACACAUAACUGUGUGUUUU-GGCCAACUGUGUGUGCGGUGGCAAUUACCAGGGGCAACAACAGGCGUCGAUGGUUCAUAAAAAUGUG-UAACGA ......((...(((....((((((((((....))))))))))-.(((..(((.((((((....)))..)))))).))).......))).))....(((((((....))))-.))).. ( -31.80) >consensus GU___AGAAAACCCAUAAAGA__________CUUGGCUUUAUGAGCUACCUGUAUGU__GGUAGCAAUUACUUCAGGCAAAAACAAACGAAGAU____UAUAAAAA______AAACA .............(((((((................))))))).((((((.........)))))).................................................... ( -8.90 = -9.12 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:14 2006