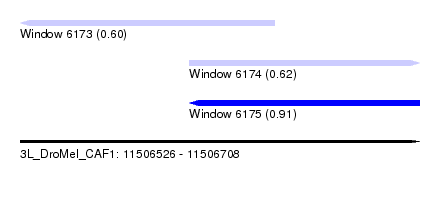
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,506,526 – 11,506,708 |
| Length | 182 |
| Max. P | 0.906377 |

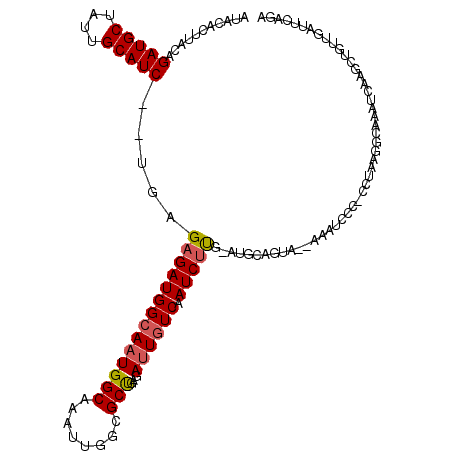
| Location | 11,506,526 – 11,506,642 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11506526 116 - 23771897 AUACACUUACAGAUGCUAUUGCAUCUUUGAGAGAUGGAAAUGGCAAAUUGGCGCUGAGAUUGUCAAUCUCGGAUGCAGUG--AAAUCUG-CCUAUGGCAAAUCUAGCUCUUGAUUCAGA ..........((((((....))))))(..((((.((((..((.((........(((((((.....)))))))..((((..--....)))-)...)).))..)))).))))..)...... ( -33.10) >DroSec_CAF1 12550 113 - 1 AUACACUUUCAGAUGCUAUUGCAUC--UGAGAGAUGGCAUUGGCAAAUUGGCGCUGAGAUUGUCAAUCUUG-AUGCAGUG--AAAUCCC-CCUAGGGCACAUCAAGCUAUUGAUUCAGA ...(((((((((((((....)))))--)))))).)).((.((((......(((..(((((.....))))).-.))).(((--....(((-....)))))).....)))).))....... ( -36.00) >DroSim_CAF1 10915 94 - 1 AUACACUUUCAGAUGCUAUUGCAUC--UGAGAGAUGGCAAUGGCAAAUUGGCGCUGAGAUUGUCAAUCUUG-AUGCA----------------------AAUCAACAUUUUAAUUCAGA .....(((((((((((....)))))--))))))..(.((((.....)))).).(((((.(((..(((.(((-((...----------------------.))))).))).)))))))). ( -28.70) >DroEre_CAF1 6809 114 - 1 AUACAUUUCCCGAUGCUAUUGCAUC--UCAGAGAUGGCAAUGGCAAAUUGGCGCCGAGAUUGUCAAUCUUG-AUACAGUA--AAGUCCCCCUUAAGGCAAAUCAAACUGUUGGUCCGGA .........(((.((((((((((((--.....))).)))))))))....(((..((((((.....))))))-..(((((.--.......((....))........)))))..)))))). ( -28.89) >DroYak_CAF1 10910 116 - 1 AUACAUUUACAGAUGCUGCUGCAUC--UGAGUGAUGGCAAUGGCAAAUUGGCGCCGAGAUUGUCAAUCUUG-AUACAGUACAAAUCCCCCCCUAAGGCAAAUCGAGCUGUUGUUCCAGA ...((((..(((((((....)))))--)))))).((((((((((........)))...))))))).(((.(-(.(((((........((......))........)))))...)).))) ( -28.69) >consensus AUACACUUACAGAUGCUAUUGCAUC__UGAGAGAUGGCAAUGGCAAAUUGGCGCUGAGAUUGUCAAUCUUG_AUGCAGUA__AAAUCCC_CCUAAGGCAAAUCAAGCUGUUGAUUCAGA ...........(((((....))))).....((((((((((((((........)))...)))))).)))))................................................. (-15.84 = -16.04 + 0.20)

| Location | 11,506,603 – 11,506,708 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11506603 105 + 23771897 UUCCAUCUCUCAAAGAUGCAAUAGCAUCUGUAAGUGUAUCUGCAUCUGGAAACCGGUUGGAACCGUUCACCCGCCAGAUAUU------CAUACAGAAUGCAU------AAUAGCGAA ...(((((.....))))).....(((((((((.(.(((((((.....(....)(((.((((....)))).))).))))))).------).))))).))))..------......... ( -27.60) >DroSec_CAF1 12626 115 + 1 UGCCAUCUCUCA--GAUGCAAUAGCAUCUGAAAGUGUAUCUGCAUCUGAAAACCGGUUGGAACCGUUCACCCGCCAGAUAUUCAUAUUCAUACAGAAUGCAUAAUCAUAAUAGCGGA (((...((.(((--(((((....)))))))).)).)))((((((((((.....(((.((((....)))).))).))))).....(((((.....))))).............))))) ( -29.90) >DroSim_CAF1 10972 109 + 1 UGCCAUCUCUCA--GAUGCAAUAGCAUCUGAAAGUGUAUCUGCAUCUGAAAACCGGUUGGAACCGUUCACCCGCCAGAUAUUCAUAUUCAUACAGAAUGCAU------AAUAGCGGA (((...((.(((--(((((....)))))))).)).)))((((((((((.....(((.((((....)))).))).))))).....(((((.....)))))...------....))))) ( -29.90) >DroEre_CAF1 6886 102 + 1 UGCCAUCUCUGA--GAUGCAAUAGCAUCGGGAAAUGUAUCUGCAUCUGG-AACCGGUUGGAACCGUUCACCCGCCAGAU------AUUCAUACAGAAUGCAU------AAUAGCGGA ...(((.(((..--(((((....))))).))).)))..(((((((((((-...(((.((((....)))).)))))))))------((((.....))))....------....))))) ( -29.80) >DroYak_CAF1 10989 104 + 1 UGCCAUCACUCA--GAUGCAGCAGCAUCUGUAAAUGUAUCUGCAUCUGA-AACCGGUUGGAACCGUUCACCCGCCAGAUA----UAUUCAUACAGAAUGCAU------AAUAGCGGA .........(((--(((((((..((((......))))..))))))))))-...((((....)))).....((((...((.----(((((.....))))).))------....)))). ( -30.60) >consensus UGCCAUCUCUCA__GAUGCAAUAGCAUCUGAAAGUGUAUCUGCAUCUGAAAACCGGUUGGAACCGUUCACCCGCCAGAUAUU__UAUUCAUACAGAAUGCAU______AAUAGCGGA ..((..........(((((....))))).....(((((((((.(((((.....(((.((((....)))).))).))))).............))).))))))............)). (-19.46 = -19.42 + -0.04)

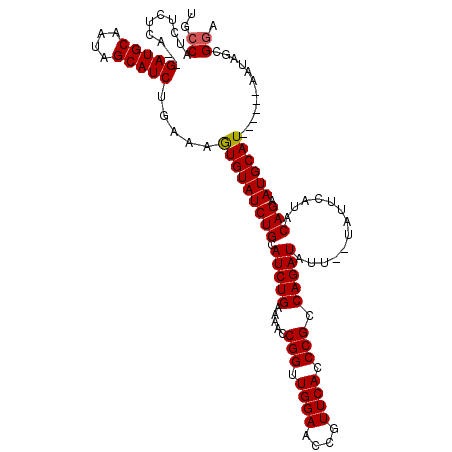
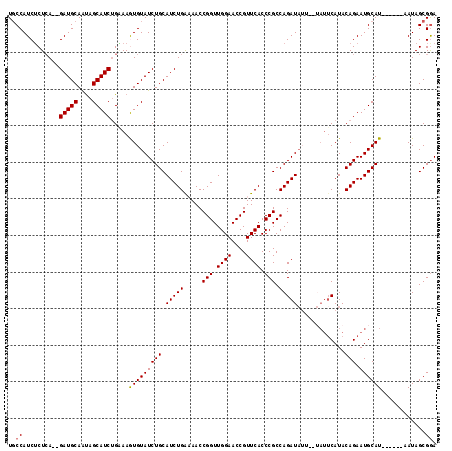
| Location | 11,506,603 – 11,506,708 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.61 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
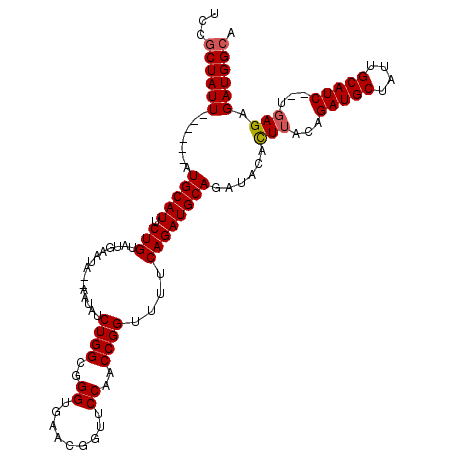
>3L_DroMel_CAF1 11506603 105 - 23771897 UUCGCUAUU------AUGCAUUCUGUAUG------AAUAUCUGGCGGGUGAACGGUUCCAACCGGUUUCCAGAUGCAGAUACACUUACAGAUGCUAUUGCAUCUUUGAGAGAUGGAA ....(((((------.(((((.(((...(------((...((((..((.........))..)))).)))))))))))......((((.((((((....)))))).)))).))))).. ( -30.60) >DroSec_CAF1 12626 115 - 1 UCCGCUAUUAUGAUUAUGCAUUCUGUAUGAAUAUGAAUAUCUGGCGGGUGAACGGUUCCAACCGGUUUUCAGAUGCAGAUACACUUUCAGAUGCUAUUGCAUC--UGAGAGAUGGCA ...(((((((((.((((((.....)))))).))))).((((((.((..((((((((....))))...))))..))))))))..(((((((((((....)))))--)))))).)))). ( -41.70) >DroSim_CAF1 10972 109 - 1 UCCGCUAUU------AUGCAUUCUGUAUGAAUAUGAAUAUCUGGCGGGUGAACGGUUCCAACCGGUUUUCAGAUGCAGAUACACUUUCAGAUGCUAUUGCAUC--UGAGAGAUGGCA ...(((((.------......(((((((.....((((...((((..((.........))..))))..)))).)))))))....(((((((((((....)))))--))))))))))). ( -39.50) >DroEre_CAF1 6886 102 - 1 UCCGCUAUU------AUGCAUUCUGUAUGAAU------AUCUGGCGGGUGAACGGUUCCAACCGGUU-CCAGAUGCAGAUACAUUUCCCGAUGCUAUUGCAUC--UCAGAGAUGGCA ...((..((------((((.....)))))).(------(((((.((.(.(((((((....))).)))-))...))))))))((((((..(((((....)))))--...)))))))). ( -30.60) >DroYak_CAF1 10989 104 - 1 UCCGCUAUU------AUGCAUUCUGUAUGAAUA----UAUCUGGCGGGUGAACGGUUCCAACCGGUU-UCAGAUGCAGAUACAUUUACAGAUGCUGCUGCAUC--UGAGUGAUGGCA ...((((((------((..........(((((.----((((((.((..(((.((((....))))...-)))..)))))))).)))))(((((((....)))))--)).)))))))). ( -37.30) >consensus UCCGCUAUU______AUGCAUUCUGUAUGAAUA__AAUAUCUGGCGGGUGAACGGUUCCAACCGGUUUUCAGAUGCAGAUACACUUACAGAUGCUAUUGCAUC__UGAGAGAUGGCA ...((((((.......(((((.(((...............((((..((.........))..))))....))))))))......(((...(((((....)))))...))).)))))). (-24.45 = -24.61 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:10 2006