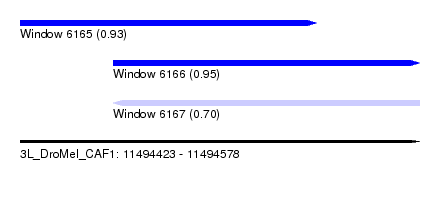
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,494,423 – 11,494,578 |
| Length | 155 |
| Max. P | 0.945685 |

| Location | 11,494,423 – 11,494,538 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
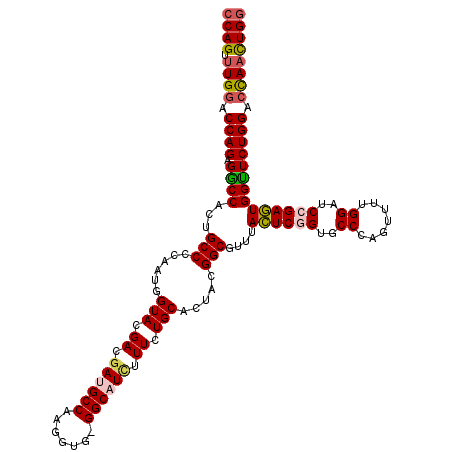
>3L_DroMel_CAF1 11494423 115 + 23771897 UUUAUUUCGGAUUGCUAAU---CAAACAUUCAUACUUU-UCCAGUUGGUCCAGAACCACUCGGAACCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCA ((((((.(((..(((....---...............(-((((((.(((.....)))))).))))(((....))))))))))))))).((((..(((((......))).-.))..)))). ( -27.50) >DroSec_CAF1 9515 103 + 1 ------------UGCCAAU---CAAGCAUUCAUACUUU-UCCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCA ------------.(((((.---...(((((........-.(((((((((((((.....)).)))))...))))))((((((.((....))))..)))).....))))).-....))))). ( -29.50) >DroSim_CAF1 4248 103 + 1 ------------UGCCAAU---CAAGCAUUCAUACUUU-UCCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCA ------------.(((((.---...(((((........-.(((((((((((((.....)).)))))...))))))((((((.((....))))..)))).....))))).-....))))). ( -29.50) >DroPer_CAF1 8752 117 + 1 NNNNNNNNNNNNNNNNNNN---NNNNNNNNNUCACUUAACCCAAUUAUGCCAGAGCCAUUCGGAGCCGAAGCUCGGCACCGAAUAUACGCCGUAGUGCAAAAAAAUGCCCCACUUUGGCC ...................---..........................(((((((..((((((.((((.....)))).))))))............(((......)))....))))))). ( -26.90) >DroEre_CAF1 5411 113 + 1 U-----AUCGAUUCACAUUCUUCGAACAUUCAUACUUU-UCCAGUUGGUCCAGAACCACUCGGAUCCGAAACUGGGCACAGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCA .-----.((((..........)))).......((((((-((((((((((((((.....)).)))))...)))))))...))))))...((((..(((((......))).-.))..)))). ( -28.60) >DroYak_CAF1 6019 110 + 1 U-----UUCGAUUGCCAAU---CAGGCAUUCAUACUUU-UCCAGUUAGUCCAGAACCACUCGGAUCCAAAACUGGGCACUGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCC .-----.......(((((.---..((((((........-.((((((.((((((.....)).))))....))))))((((((.((....))..)))))).....))))))-....))))). ( -31.20) >consensus ____________UGCCAAU___CAAGCAUUCAUACUUU_UCCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC_CACCUUGGCA ................................(((((..((((((((((((.((.....)))))))...)))))))....)))))...((((..(((((......)))...))..)))). (-19.97 = -20.80 + 0.83)


| Location | 11,494,459 – 11,494,578 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.11 |
| Mean single sequence MFE | -49.35 |
| Consensus MFE | -39.97 |
| Energy contribution | -40.95 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11494459 119 + 23771897 CCAGUUGGUCCAGAACCACUCGGAACCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCAUCGUCGUACCAUUGGGGCAAUGGUCUCUGGUCCAAACUGG ((((((((.((((((((((((((..(((....)))...))))))....(((.(((((..((..((((((-......)))))).))....))))).)))...))).))))).)).)))))) ( -55.00) >DroSec_CAF1 9539 119 + 1 CCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCAUCGUCGUACCAUUGGGGCAGUGGCCUCUGGUCCAAACUGG ((((((((.(((((.((((((((.((((....))))..))))))....(((.(((((..((..((((((-......)))))).))....))))).)))...))..))))).)).)))))) ( -53.00) >DroSim_CAF1 4272 119 + 1 CCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCAUUGUCGUACCAUUGGGGCAGUGGCCUCUGGUCCAAACUGG ((((((((.(((((.((((((((.((((....))))..))))))....(((.(((((..((..((((((-......)))))).))....))))).)))...))..))))).)).)))))) ( -51.20) >DroPer_CAF1 8789 120 + 1 CCAAUUAUGCCAGAGCCAUUCGGAGCCGAAGCUCGGCACCGAAUAUACGCCGUAGUGCAAAAAAAUGCCCCACUUUGGCCUCAUCAUACCAACGGGGCAGGGGCCGCUGGUCCAGACUGG (((.((..(((((.(((((((((.((((.....)))).)))))).....(((.....).......((((((...((((..........)))).)))))))))))..)))))...)).))) ( -43.50) >DroEre_CAF1 5445 119 + 1 CCAGUUGGUCCAGAACCACUCGGAUCCGAAACUGGGCACAGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCAUCGUCGUACCACUGGGGCAGUGGUCUCUGGUCCAAGCUGC .(((((((.(((((((((((((....)))..(((....))).......(((.(((((..((..((((((-......)))))).))....))))).))).))))).))))).)).))))). ( -51.40) >DroYak_CAF1 6050 119 + 1 CCAGUUAGUCCAGAACCACUCGGAUCCAAAACUGGGCACUGAGUAAACGCCGUAGUGCAGAAAGAUGCC-CACCUUGGCCUCGUCGUACCAUUGGGGCAGUGGCCUCUGGUCCAAGCUGG ((((((.(.(((((.((((((((.((((....))))..))))))....(((.(((((..((..((.(((-......))).)).))....))))).)))...))..))))).)..)))))) ( -42.00) >consensus CCAGUUGGUCCAGAACCACUCGGAUCCAAAACUGGGCACCGAGUAAACGCCGUAGUGCAGAAAGAUGCC_CACCUUGGCAUCGUCGUACCAUUGGGGCAGUGGCCUCUGGUCCAAACUGG ((((((((.(((((.((((((((..(((....)))...))))))....(((.(((((......((((((.......)))))).......))))).)))...))..))))).)).)))))) (-39.97 = -40.95 + 0.98)

| Location | 11,494,459 – 11,494,578 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.11 |
| Mean single sequence MFE | -49.64 |
| Consensus MFE | -39.72 |
| Energy contribution | -39.95 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
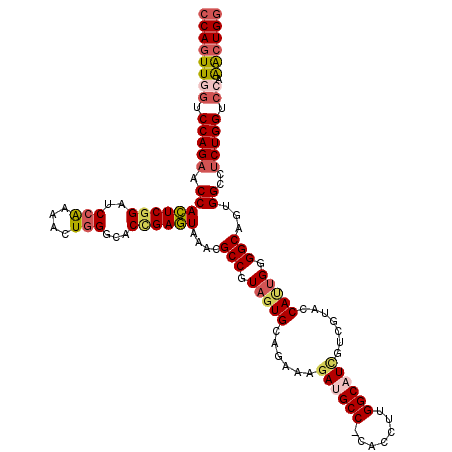
>3L_DroMel_CAF1 11494459 119 - 23771897 CCAGUUUGGACCAGAGACCAUUGCCCCAAUGGUACGACGAUGCCAAGGUG-GGCAUCUUUCUGCACUACGGCGUUUACUCGGUGCCCAGUUUUGGUUCCGAGUGGUUCUGGACCAACUGG ((((((.((.((((((.((((((((.....)))....((..(((((((((-((((((.....((.(....).))......)))))))).)))))))..))))))))))))).)))))))) ( -51.50) >DroSec_CAF1 9539 119 - 1 CCAGUUUGGACCAGAGGCCACUGCCCCAAUGGUACGACGAUGCCAAGGUG-GGCAUCUUUCUGCACUACGGCGUUUACUCGGUGCCCAGUUUUGGAUCCGAGUGGUUCUGGACCAACUGG ((((((.((.((((((.((((((((.....))).((..(((.((((((((-((((((.....((.(....).))......)))))))).)))))))))))))))))))))).)))))))) ( -51.60) >DroSim_CAF1 4272 119 - 1 CCAGUUUGGACCAGAGGCCACUGCCCCAAUGGUACGACAAUGCCAAGGUG-GGCAUCUUUCUGCACUACGGCGUUUACUCGGUGCCCAGUUUUGGAUCCGAGUGGUUCUGGACCAACUGG ((((((.((.((((((.((((((((.....))).........((((((((-((((((.....((.(....).))......)))))))).)))))).....))))))))))).)))))))) ( -49.60) >DroPer_CAF1 8789 120 - 1 CCAGUCUGGACCAGCGGCCCCUGCCCCGUUGGUAUGAUGAGGCCAAAGUGGGGCAUUUUUUUGCACUACGGCGUAUAUUCGGUGCCGAGCUUCGGCUCCGAAUGGCUCUGGCAUAAUUGG (((((...(.((((.((((..((((((.(((((........)))))...))))))......(((.(....).))).((((((.((((.....)))).)))))))))))))))...))))) ( -51.60) >DroEre_CAF1 5445 119 - 1 GCAGCUUGGACCAGAGACCACUGCCCCAGUGGUACGACGAUGCCAAGGUG-GGCAUCUUUCUGCACUACGGCGUUUACUCUGUGCCCAGUUUCGGAUCCGAGUGGUUCUGGACCAACUGG .(((.((((.((((((.((((((((((..(((((......)))))..).)-)))((((..(((...(((((.(....).)))))..)))....))))...))))))))))).))))))). ( -46.20) >DroYak_CAF1 6050 119 - 1 CCAGCUUGGACCAGAGGCCACUGCCCCAAUGGUACGACGAGGCCAAGGUG-GGCAUCUUUCUGCACUACGGCGUUUACUCAGUGCCCAGUUUUGGAUCCGAGUGGUUCUGGACUAACUGG ((((.((((.((((((.((((((((.....)))....((.((((((((((-(((((......((.(....).)).......))))))).)))))).))))))))))))))).)))))))) ( -47.32) >consensus CCAGUUUGGACCAGAGGCCACUGCCCCAAUGGUACGACGAUGCCAAGGUG_GGCAUCUUUCUGCACUACGGCGUUUACUCGGUGCCCAGUUUUGGAUCCGAGUGGUUCUGGACCAACUGG ((((.((((.((((.((((...(((......(((.((.((((((.......)))))).)).))).....)))....((((((..((.......))..)))))))))))))).)))))))) (-39.72 = -39.95 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:02 2006