| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,493,906 – 11,494,014 |
| Length | 108 |
| Max. P | 0.975724 |

| Location | 11,493,906 – 11,494,014 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11493906 108 - 23771897 ACAUGUAUACAUU----CU--U-----AUUUUCUAUUCUC-UUAGAGGAACUGGCUGCCGCAAUUCGAAAGGAGUCCGAUCUUCGUUUUGGACUCUAUUAUUCUCUAUUCGAAUGGUUCA .............----..--.-----.....((((((..-.(((((((....((....)).........(((((((((........)))))))))....)))))))...)))))).... ( -27.00) >DroSec_CAF1 8960 108 - 1 ACGUAUAUACAUU----UA--U-----AUUUUCUACUCUC-CCAGAGGAACUGGCUGCCGCAAUUCGAAAGGAGUCCGAUCUUCGUUUCGGACUCUACUAUUCUUUGUUUGAAUGGUUCA ..(((((......----))--)-----))((((..(((..-...)))(((...((....))..)))))))(((((((((........)))))))))(((((((.......)))))))... ( -27.90) >DroSim_CAF1 3688 108 - 1 ACGUAUAUACAUU----CU--U-----AUUUUCUAUUCUC-CCAGAGGAACUGGCUGCCGCAAUUCGAAAGGAGUCCGAUCUUCGUUUCGGACUCUACUAUUCUCUGUUCGAAUGGUUCA .............----..--.-----.....((((((..-.(((((((....((....)).........(((((((((........)))))))))....)))))))...)))))).... ( -29.90) >DroPer_CAF1 8189 111 - 1 GGAGGCAUAGAUU----CAGAU-----AUAUACCGCCCUCCACAGAGGAACUUGCGGCGGCCGUUCGCGGUGUUUCGGAUCUGAAGUUCGGCCUGUACUAUUCCCUCUUCGAGUGGUUCA ..((((.....((----(((((-----..(((((((((((....)))).....((((...))))..))))))).....))))))).....))))(.(((((((.......))))))).). ( -38.50) >DroEre_CAF1 4644 108 - 1 ---------AAUUAUAUGU--UAAUACACUUUGCAUUUUC-CCAGAGGAACUGGCUGCCGCUAUACGAAAGGAGUCCGAUCUUCGGUUCGGACUUUACUAUUCCCUAUUCGAAUGGUUCA ---------..........--.......(((((.......-.)))))((((((((....)))...((((((((((((((.(....).))))))).........))).))))...))))). ( -27.30) >DroYak_CAF1 5387 103 - 1 ---------AAUU----CU--UAACAAAAUU-CAAUUUUC-CCAGAGGAACUGGCCGCCGCCAUUCGAAAGGAGACCAAUCUUCGGUUUGGACUCUACUACUCCCUAUUCGAGUGGUUCA ---------....----..--........((-(....(((-(....)))).((((....))))...))).((((.((((.(....).)))).))))(((((((.......)))))))... ( -25.40) >consensus AC_U_UAUACAUU____CU__U_____AUUUUCUAUUCUC_CCAGAGGAACUGGCUGCCGCAAUUCGAAAGGAGUCCGAUCUUCGGUUCGGACUCUACUAUUCCCUAUUCGAAUGGUUCA ...............................................(((((.((....)).(((((((.(((((((((........)))))))))...........)))))))))))). (-17.11 = -17.50 + 0.39)

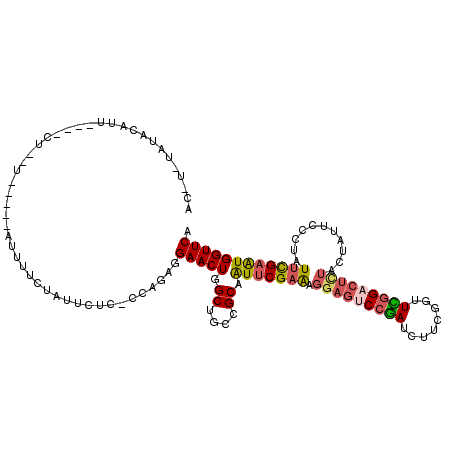

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:59 2006